Population Biology: The Epidemiology and Population Genetics of Plant Pathogens
A population is a group of organisms from the same species that occupies the same geographic region and exhibits reproductive continuity from generation to generation. We usually assume that ecological and reproductive interactions are more common among members of the same population than among members from different populations.
Epidemiology and population genetics are different, but related, subsets of population biology. Plant disease epidemiology is the study of the distribution and determinants of disease frequency in plant populations. Epidemiology focuses on disease progression, the multiplication of pathogen populations through time and the movement of pathogen populations through space (usually from plant to plant). The majority of epidemiological studies deal with short time scales (e.g. 1-2 growing seasons) and small spatial scales (e.g. disease development within a field or a plantation), though some epidemiological studies include analyses across continents. Epidemiology involves mainly physical processes such as distance of spore movement or effects of weather variables on latent periods. Epidemiology usually does not take into account differences in the behavior or fitness of genetically distinct individuals in a collection of individuals.
Population genetics focuses on the processes that lead to genetic change, or evolution, in populations over time and space. The majority of population genetics studies deal with longer time scales (e.g. multiple growing seasons, tens to hundreds of pathogen generations) and larger spatial scales (e.g. often global for pathogens with spores that move tens or hundreds of kilometers). Population genetics deals mainly with genetic processes, such as mutation, genetic drift, gene flow, mating systems, and natural selection.
Factors Shared by Epidemiology and Population Genetics
There is considerable overlap between population genetics and epidemiology, as shown in Figure 1. One key element for both is the dispersal distance of pathogen propagules, which is important for measuring initial inoculum load in epidemiology and important for measuring gene/genotype flow in population genetics. Another unifying element is the latent period, which affects the shape of the disease progress curve in epidemiology, and is a fitness parameter in population genetics. A third element is the size of the pathogen population, which affects the measure of disease incidence in epidemiology and the degree of genetic drift in population genetics. Another major feature that is shared by population genetics and epidemiology is the importance of time.
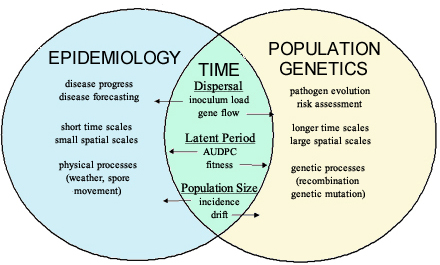
Figure 1. Epidemiology and population genetics are related subsets of population biology with many overlapping concepts, but employing different approaches. AUDPC = area under the disease progress curve.
Time is an important component of epidemiology. In addition to a susceptible host, a virulent pathogen, and a suitable environment (the components of the disease triangle), time is needed for pathogens to complete their life cycle, produce offspring, and increase the size of the pathogen population. An increase in population size is followed by a new cycle of infection and disease development, adding another step in the epidemic cycle. Time also is important for population genetics, because time is needed for populations to evolve.
What is Evolution?
Evolution is a two-step process. First, we have processes such as mutation that give rise to genetic variation in populations. Then processes like selection or genetic drift act to change allele frequencies in populations. Evolution results from changes in allele frequencies in populations. As an example, when a susceptible plant population experiences an increase in the frequency of a resistance allele, resulting from selection for resistant individuals, the plant population has evolved to a higher level of disease resistance. And when an avirulent pathogen population experiences an increase in the frequency of a virulence allele as a result of selection for virulent individuals that can evade recognition by a plant resistance allele, the pathogen population has evolved to a higher level of virulence.
Evolutionary Biology: Population Genetics and Phylogenetics in Plant Pathology
Evolutionary biology is a discipline that considers processes that lead to genetic change, or evolution, in populations or species of organisms. Population genetics and phylogenetics are subdisciplines within evolutionary biology (Figure 2). Population genetics considers microevolutionary processes that occur within species and attempts to explain the distribution of genetic variation within and among populations of a single species. Phylogenetics considers macroevolutionary processes that occur among species and attempts to explain the lines of ancestry that led to the current distribution of species across time and space.

Figure 2. The connection between phylogenetics and population genetics lies in the process of speciation. Population genetics considers mainly processes that affect population while phylogenetics considers mainly processes that affect species.
Evolutionary biology is relevant to plant pathology because pathogen populations evolve in response to control methods and it often is not clear whether a new pathogen population is derived from a pre-existing pathogen population or represents a host-jump or a new species of pathogen. Phylogenetics classifies species based on lines of descent from common ancestors, while population genetics determines relationships among populations within a species. The interface between population genetics and phylogenetics is in the emergence of new species, a process called speciation. Speciation may occur more rapidly for plant pathogens than for other organisms as a result of coevolution.
Phylogenetics and population genetics use different genetic and analytical tools. Phylogenetic analysis is based on using species-specific characters to define taxonomic units based on the methods of cladistics or phenetics. Population genetics is based on using allele frequencies at polymorphic loci to determine genetic structure and define population boundaries. Both areas of research try to elucidate the historical processes and natural history characteristics that led to the current composition of populations and species. Population genetics and phylogenetics often overlap in the fields of biogeography and phylogeography.
When trying to determine the origins of new pathogens, plant pathologists may end up using tools of both phylogenetics and population genetics.
Coevolution Between Plants and Pathogens
When considering plant pathogens, many evolutionary biologists think of plant disease in natural ecosystems as a coevolutionary process. Coevolution is a process during which a trait of one species evolves in response to a trait of another species, which trait in turn evolves in response to the trait in the first species (Janzen 1980). Coevolution studies consider the reciprocal genetic changes that are expected to occur in two or more ecologically interacting species. The fundamental element of coevolution is reciprocity. A genetic change in one species causes a genetic change in the coevolving species. And this genetic change in turn causes new genetic change in the first species. Reciprocity appears to occur in natural plant pathosystems as illustrated in Figure 3.

Figure 3. Coevolution in a natural plant pathosystem is characterized by the development of resistance in the host and virulence in the pathogen. The two characters are interconnected.
This coevolution process is thought to occur in natural ecosystems where plant and pathogen exhibit a gene-for-gene (GFG) relationship, as summarized in the quadratic check shown below:
|
Quadratic check in GFG systems |
| |
Host |
|
Pathogen |
R_ (resistant) |
rr (susceptible) |
|
A (avirulent) |
- resistant |
+ susceptible |
|
a (virulent) |
+ susceptible |
+ susceptible |
According to our present knowledge of GFG interactions, the avirulence allele encodes an elicitor that is recognized by a specific receptor (encoded by the R-allele) in the plant host (Figure 4). Recognition of the elicitor by the receptor initiates a signal-transduction pathway that turns on genes involved in the hypersensitive response.
 |
| Figure 4. The receptor-elicitor model of gene-for-gene interactions. The resistance allele of the plant encodes a receptor that recognizes an elicitor produced by the pathogen. Recognition of the pathogen elicitor by the plant receptor initiates plant defense responses that lead to plant resistance. If the pathogen produces the elicitor, it is avirulent. If the pathogen does not produce the elicitor, it is virulent. Click here to see an enlarged view of this figure. |
The Logic of GFG Coevolution in Natural Ecosystems
Coevolution begins with a plant population affected by a disease. Within the diseased plant population, a mutation or recombination event occurs that produces a novel receptor allele that recognizes an elicitor in the pathogen. This event causes the mutation from susceptibility to resistance in the plant. In the presence of constant disease pressure that reduces the fitness of susceptible plants, the resistant plants will produce more offspring and thus the resistance allele will increase in frequency. As the resistance in the plant population increases due to an increase in the frequency of resistant individuals, the ability of the pathogen to reproduce is reduced and the overall level of disease decreases. At this point, there is selection for individuals in the pathogen population that have a mutation causing the loss of the avirulence allele (the elicitor) or a mutation in the avirulence allele that alters the shape of the elicitor so that it is no longer recognized by the plant resistance allele (receptor). This is the mutation from avirulence to virulence in the pathogen. If the pathogen population has a mutation from avirulence to virulence, there will be selection that favors the virulence allele in the resistant plant population. Once the mutation to virulence occurs, the frequency of the virulence allele increases, leading to an increase in the amount of disease in the previously resistant plant population. And the cycle begins again. Many theories predict that plant alleles for resistance and susceptibility, and pathogen alleles for virulence and avirulence should coexist in natural populations for a long time. Theories also predict that coevolution will occur at the fastest pace in places where the amount of disease is greatest.
Several examples of coevolution exist in natural plant pathosystems. Gene-for-gene interactions between plant resistance genes and pathogen avirulence genes have been demonstrated mainly for obligate biotrophs such as rust fungi and powdery mildew fungi in wild cereal populations. Examples include the wild flax-flax rust fungus populations in Australia (Jeremy J. Burdon and colleagues), and wild oat and oat rust fungus populations in Israel (Isaac Wahl and colleagues).
Human-Guided Evolution in Agroecosystems
The reciprocity that is a hallmark of coevolution does not truly exist in agricultural ecosystems. In agriculture, humans control the plant part of the interaction, by determining which plant genotypes are grown in a farmer's field. The pathogen evolves in response to the resistance alleles that are present in the plant, but the plant population is not given a chance to respond naturally to the changes in the frequency of virulence alleles that occur in the pathogen population. When a resistance gene is overcome, plant breeders replace it with another resistance gene, creating the boom and bust cycle. Plant and pathogen populations are not given a chance to reach a natural equilibrium where resistance alleles and virulence alleles coexist for long periods of time. Thus we can say that coevolution does not really occur in agricultural systems. Rather, the evolution of the plant pathogen is directed by humans, and in particular by plant breeders.
In agroecosystems we are concerned with how a control method (e.g. introduction of a resistance gene, application of a fungicide, initiating a quarantine, crop rotation, etc.) affects the population genetics, or evolution, of the targeted pathogen population.
In population genetics, we generally consider five evolutionary forces that affect pathogen populations. These forces are mutation, genetic drift, reproduction and mating system, gene flow, and natural selection. In the absence of the effects of mutation, genetic drift, gene flow, and selection, we expect that populations will be at Hardy-Weinberg equilibrium.
For simplicity, we will begin by considering only one evolutionary force at a time. But in nature, all of these forces interact to determine the course of evolution in a pathogen population and to generate the genetic structure of pathogen populations. We will begin with a short overview that considers each of the forces individually.
Mutation is a change in the DNA at a particular locus in an organism. Mutation is the ultimate source of new alleles in plant pathogen populations. It also is the source of new alleles that create new genotypes within clonal lineages. Small populations have fewer alleles due to genetic drift and because there are fewer individuals in which mutations can be generated. Old populations have more neutral alleles than new populations, assuming that population sizes are equal in new and old populations.
Genetic drift is a random process that can lead to unpredictable changes in pathogen populations over a short period of time. Random drift is caused by recurring small population sizes, severe reductions in population size called "bottlenecks" and founder events where a new population starts from a small number of individuals. Genetic drift leads to fixation of alleles or genotypes in populations and therefore tends to decrease overall levels of genetic variation.
Gene flow breaks down the boundaries that could otherwise isolate populations. Gene flow is especially important for plant pathogens in agroecosystems because it is the process that introduces new genetic variation into agricultural fields distant from the site of the original mutation.
Reproductive/mating systems affect the way that alleles are put together in each individual in a population. Outcrossing pathogens put together new combinations of alleles rapidly, leading to many different genotypes in populations. Pathogens that are inbreeding or that undergo asexual reproduction tend to keep together existing combinations of genes leading to lower genotype diversity in populations. Pathogens that have "mixed" mating/reproduction systems, including both sexual and asexual reproduction, potentially benefit from the advantages inherent to both types of reproduction.
Selection is a directional process that leads to an increase in the frequency of selected alleles or genotypes. Selection is the process that increases the frequencies of plant resistance alleles in natural ecosystems through coevolution, and it is the process that increases the frequencies of virulence alleles in agricultural ecosystems during boom and bust cycles.
Go to Knowledge Test for Interactions/Genetic Structure
Go to References
Next Section
