Introduction
| |
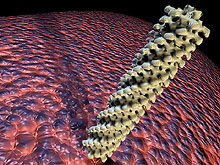
Fig. 1. The contractile tail sheath of phage T4 of Escherichia coli, one of the best understood phages. This computer-generated image is based on the data of J. Lepault and K. Leonard (46) and was generated by Steven McQuinn. |
Plant biology cannot be fully appreciated absent microbial flora, and plant-associated bacteria are incompletely understood without an awareness of phage -- the viruses of prokaryotes. Phage have been found in association with “buds, leaves, root nodules (leguminous plants), roots, rotting fruit, seeds, stems and straw; crown gall tumors… healthy or diseased alfalfa, barley, beans, broccoli, Brussels sprouts, buckwheat, clover, cotton, cucumber, lucerne, mulberry, oats peas, peach trees, radish, rutabaga, ryegrass, rye, timothy, tobacco, tomatoes, [and] wheat” (4). In this overview we consider the myriad ways that phage can impact ecologically on plant-associated bacteria.
Table of Contents
Introduction to Phage Biology
Phage Ecology
Phage Temperance versus Virulence
Ecology of Lysogeny
Lysogeny in the Face of Plants
The Phage Rhizosphere
The Phage Phyllosphere
Phage and Agriculture
Phage Therapy
Introduction to Phage Biology
Much is known of phage biology, particularly at the molecular level (14,35). Older reviews of the biology of various phage of plant pathogens are also found elsewhere (16,52). We begin this review by focusing on the phage biology, starting with the phage life cycle, which consists of an extracellular search, attachment to susceptible bacteria, phage-genome uptake into bacteria, production (maturation) of phage progeny, and subsequent release of these progeny into the extracellular environment.
The phage itself (Fig. 2), the virion particle, consists of a nucleic acid core that is made up, depending on the phage, of DNA or, less often, of RNA. Surrounding this nucleic-acid genome is a protein-based capsid. The capsid plays three important roles in the phage life cycle: (i) protecting the phage genome during the extracellular search (e.g., from DNA-degrading enzymes); (ii) effecting phage adsorption, which is the attachment of the virion particle to a susceptible bacterium; and (iii) the subsequent delivery (uptake) of the phage genome into the cytoplasm of the now-infected bacterium.
| |
 |
|
| |
Fig. 2. Electron micrograph of phage T2 courtesy of Hans-Wolfgang Ackermann. Note the prominent tail sheath running like a slightly bent cylinder from just beneath the phage head to the phase baseplate found towards the bottom of the image. Note also the six long tail fibers faintly visible and attached to the baseplate. The tail fibers are the primary adsorption organelles for this phage. The baseplate irreversibly attaches the capsid to the cell surface. Tail sheath contraction punctures the enclosed tail tube through the cell envelope. Phage DNA, contained within the phage head, subsequently is transported through the tail tube and into the now-infected bacterium's cytoplasm. |
|
The extracellular search occurs via phage diffusion through an aqueous milieu. During this period the phage must avoid physical damage while waiting to encounter a susceptible bacterium. The likelihood that an individual phage will find a bacterium to adsorb is a function of time, the phage diffusion rate, and the local density of phage-susceptible bacteria, with more bacteria resulting in faster phage adsorption (36). A slightly different set of parameters governs the likelihood of phage attack on bacteria, with adsorption a function of time and the phage diffusion rate, but also of phage density, with more phage resulting in more bacteria infected (2,36).
Phage adsorption to bacteria furthermore is a function of phage-bacteria chemical and physical interaction. Phage display proteins with high affinity to specific bacterial surface molecules-an association analogous to antigen recognition by immune systems (30). The host range of most phages, i.e., the species that they are capable of productively infecting, consequently is relatively narrow -- typically limited to only a single bacterial genus, species, or, often, even to only a limited number of strains within a given species. Thus, while total phage densities can be enormous -- as many as 100 million or more per gram of soil or ml of aquatic environment (8,82) -- the actual density of phages capable of infecting a particular bacterial strain usually is much smaller.
Following uptake, the phage genome can rapidly subvert host-cell functions, directing bacterial metabolism during the phage latent period towards phage production. Depending on the phage, these virions either may accumulate within the bacterial cytoplasm (as is the case for so-called lytic phage) or, for filamentous phage, virions instead are released -- over the course of an extended latent period -- across an otherwise intact cell envelope. Once released, one describes the virion as a free phage.
Phage Ecology
Phage ecology (e.g., 3) is the study of the interaction of phage with their biotic and abiotic environments. Much of this review considers the ecological interaction between phage and bacteria within the context of plants and their surrounding soil, and in this section we introduce the basics of phage ecology. Following the traditions of ecology we can differentiate phage ecology into four general categories: (i) Phage organismal ecology is the study of the adaptations that phage employ to increases their likelihood of transmission between hosts such as virion desiccation resistance, ability during infection to repair ultraviolet (UV) light-mediated nucleic-acid damage, and so on. (ii) Phage population ecology is the study of phage life-history characteristics, particularly as they apply to phage growth and intraspecific (between-phage) competition. Understanding phage population growth within spatially structured environments, such as within the phyllosphere or rhizosphere, is particularly challenging. (iii) Phage community ecology focuses on the stability of phage-containing environments such as the propensity of phage to drive phage-sensitive bacteria to extinction. Phage community ecology is complicated by the continuous co-evolution of bacteria with their phage predators. In addition, phage play important roles in the horizontal transfer of DNA between bacteria (e.g., 41). (iv) Phage ecosystem ecology considers the phage impact on energy flow and nutrient cycling within ecosystems. Phage, for example, can disrupt the soil bacteria responsible for nitrogen cycling.
Phage Temperance versus Virulence
Population and organismal ecologies are concerned with the adaptations that organisms employ to enhance their Darwinian fitness over the course of their life cycles. For phage, the life-cycle steps most under their control are the durability of the virion particle, the breadth of the host range, and the details of the infection strategy. In this section we take a population-ecology approach to contrasting two infection strategies: temperance versus virulence. Subsequently we consider the impact of plants on lysogeny.
For lytic phage, progeny release can occur only following the total destruction (lysis) of the host bacterial cell. One can differentiate lytic phage into two types, temperate phage and obligately lytic (a.k.a., virulent) phage. Only temperate phage can display lysogeny, an infection that stalls shortly after the introduction of the phage genome into the host cell, which then (in most cases) integrates as a prophage into the bacterial chromosome. During lysogeny phages neither produce virions nor lyse bacteria. A temperate phage does not obligately enter into a lysogenic relationship with its host bacterium; in fact many temperate phage infections result in the immediate production of phage progeny, i.e., a lytic cycle rather than lysogeny. This decision is determined by characteristics of the infecting phage and the metabolic state of the host. When not induced, a phage in the lysogenic state replicates as a giant gene complex along with the host cell’s genome.
Lysogeny typically results in bacterial resistance to infection by similar (i.e., homologous) phage. Mutant phage that are able to bypass this resistance are described as vir mutants (for virulence). One advantage that can be associated with such virulence is an ability to actively infect homologous lysogens (bacteria lysogenized by the same phage), though establishment of this Vir phenotype can require multiple phage mutations (7). Note, though, that a diversity of virulent phage exist that are not vir mutants but instead are unrelated to temperate phage. In addition to providing a safe home to the temperate-phage genome, and blocking the replication of non-virulent homologous phage, lysogeny has the potential to alter the phenotype of the host cell, a process known as phage (or lysogenic) conversion.
Ecology of Lysogeny
What advantages are bestowed upon a phage that displays a temperate lifestyle? We can describe obligately lytic phage as essentially semelparous, with the acquisition of a bacterial cell resulting in only a single reproductive episode. As such these phage are perhaps best understood as adapted to a so-called r strategy of population growth, with a life-history emphasis on rapid population increase when resources are plentiful (resources for phage being susceptible bacteria). Aiding in this strategy is their (i) avoidance of physiological tradeoffs required for the display of lysogeny, (ii) a commitment of all progeny to lytic growth (rather than some fraction to lysogen formation), or (iii) an ability to infect lysogens. These advantages, however, come with requirements for long-term survival as free phage when bacteria are less numerous, and/or dissemination to new environments to find new bacterial hosts.
Temperate phage can also display the semelparous infection strategy of virulent phage (i.e., the lytic cycle). A fraction of infections, however, will instead result in lysogeny. Lysogeny represents an iteroparity of sorts, i.e., more than one reproductive episode per lifetime, at least so long as one is willing to accept a clonally related population of lysogens as a single individual and a sporadic induction of lytic cycles within this population as consecutive reproductive events. Such a life-history approach is the more K-like strategy whereby temperate prophage, by less-rapidly killing off their bacterial resource, may more readily sustain their population size at an environment’s carrying capacity. Filamentous phage similarly display a more semelparous life-history strategy than virulent phage, with (i) a longer-term maintenance of the host infection, (ii) only a few phage progeny released over a given interval, and (iii) with multiple intervals (i.e., a long time) over which these phage progeny are released.
Though lysogeny often is framed as an adaptation to survival within relatively unstable environments, i.e., during “hard times” (23,69), one could similarly argue that lysogeny bestows a competitive advantage on phage that is useful when bacterial populations are relatively stable, i.e., not fluctuating in size. Lysogeny thusly can be an effective K strategy so long as lysogens are not being actively killed by lytic phage. Exposure to lytic phage may be less likely in well-structured environments that limit phage diffusion and in which lysogen microcolonies exist as relatively few cells, with low lysogen number minimizing the potential for temperate-phage mutation to anti-lysogen virulence. Of course, arguing that well-structured environments can favor lysogeny over virulence is nearly a restatement of the “hard times” hypothesis whereby the “long periods of dearth” (69) required for this dominance is posited to be a consequence of environmental structure (e.g., insurmountable distances between bacterial microcolonies) rather than solely of temporal variation in resource availability.
Lysogeny in the Face of Plants
Gvozdyak (27) has suggested that plants can induce bacterial lysogens (that is, cause them to initiate their lytic cycle), a strategy that plants could employ towards the elimination of bacterial pathogens (see also 20). Perhaps similarly, Sato (63) has shown that an extract of mulberry leaves could induce lysogens of Pseudomonas syringae. It may be that these induced prophage are “abandoning ship” in response to the plant’s release of antibacterial compounds, with the phage simply following an evolutionary algorithm based on the logic that it is better to take one’s chances as a free phage than to continue to infect a dying bacterium. Also consistent with a shift away from lysogeny in the lysogeny-lytic cycle balance, Menzel et al. (49) have reported a negative impact of certain plant-growth regulators and herbicides on the initial establishment of lysogeny.
For many bacteria, plant association marks not a high likelihood of plant-induced bacterium death but, instead, a period of effective bacterial growth. Given heterogeneous bacterial populations (26) it could be advantageous for temperate phage to lyse host bacteria when times are good since not only are healthy, uninfected bacteria potentially present (23), but those bacteria also could contain non-homologous prophage capable of infecting and then lysing their own uninduced lysogens. That is, more effective bacterial growth could tip an environment from conditions that disfavor phage lytic growth and favor lysogen survival (i.e., environments in which it is better, from the phage perspective, to be a bacterium) to conditions that favor lytic growth and thereby disfavor a continued display of lysogeny.
The Phage Rhizosphere
From the phage perspective much of what is of interest in understanding the phage-plant interaction has to do with the extracellular search, a province of phage organismal ecology. How exactly do phage manage to find new bacteria to infect before succumbing to the ravages of environmentally induced virion decay? Answering this question within the rhizosphere -- the region consisting of plant roots and surrounding soil -- involves examining the impact of soil structure and chemistry on the mobility and survival of free phage and their hosts.
In most crop environments, with a few notable exceptions, most of the time the soil is only partially hydrated. The lack of a continuous aqueous phase greatly complicates predictions regarding the diffusion of free phage, as for any soil colloid. This situation is complicated further by the propensity of free phage to become trapped within biofilms (70) or reversibly adsorbed to particles, such as clays (12,81), that are commonly found in the soil. This non-specific and often reversible phage adsorbtion within soils is a function of sorbent and virion surface chemistry, virion size, and pH (15). Moreover, acidic soils can permanently inactivate free phage (54,72).
We can only speculate whether these phage-soil interactions help or hinder the phage in their search for a suitable host. Since sorption to solid substrates lowers the diffusion rates of free phage, it localizes them to specific spatial regions, and thereby limits the maximum rate at which they can encounter a suitable host cell (39). On the other hand, adsorption to clays has been suggested to have a protective effect by holding phage within a hydrated environment (81). Both states are essentially in opposition only within undisturbed soil. If the soil itself is disseminated, becomes fully hydrated, or is otherwise well mixed, then associated phage, whether free or bound, may be disseminated. Soil-adsorbed phage thus could serve as a viable infectivity pool that is tapped only as bacteria grow, diffuse, or swim into the phage vicinity, or if the soil particle itself is transferred into or onto a bacterium-containing environment.
Phage can attack bacteria directly associated with plant roots. Given the close proximity of roots to soil it seems obvious that phage attack must occur via phage diffusion from either surrounding soil or from neighboring roots. Rhizosphere bacteria, however, may gain an upper hand in what likely is a constant battle between the phage predator and bacteria prey by some combination of (i) relatively low viable counts of phage capable of infecting specific bacteria (8); (ii) relatively low rates of phage diffusion within soil, particularly under drier conditions or following phage adsorption to soil (39); (iii) relatively high rates of free-phage inactivation within soil (54); and (iv) physical (or spatial) refuges that provide bacteria with a degree of physical protection from phage (65). Indeed, one can envisage phage replication as equivalent to a nuclear chain reaction with anything damping the mobility or production of the phage “neutron” serving to limit depletion of the bacterial “fuel” (54): Significant bacterial depletion occurs only once bacteria first have achieved a critical “mass” (i.e., sufficient density).
Phage researchers have significant experience handling phage within environments containing relatively low phage densities -- and in which diffusion (and mixing) is limited -- since these are the conditions under which phage growth typically occurs in solid media. Solid-media growth involves mixing a small number of phage with a large excess of bacteria, which is then poured into a thin layer over a regular plate of agar growth medium. The soft-agar in the top layer impedes phage and bacterial diffusion while the bottom agar layer maintains a relatively constant chemical and physical state for bacterial and phage growth. Within the soft agar layer, phage populations grow as plaques, which are expanding regions of phage-induced bacterial lysis (Fig. 3), each originating from a single phage infection. Plaques appear transparent or translucent against the background of the typically more-opaque bacterial lawn growing within the agar-based substrate. Of particular relevance to understanding host-phage ecology within the rhizosphere therefore is determination of the degree to which phage growth in soil systems approximates the better understood solid-phase phage growth in laboratory media. Though within agar plaque-development theory as well as techniques for plaque-growth quantification have been fairly well developed (34,40,44,84,85,86), to our knowledge similarly fine-scale investigation has not been attempted within a soil-based medium.
| |

Fig. 3. Phage plaques on a Erwinia lawn. |
|
Individual soils likely vary spatially and temporally with regard to plaque-like growth-particularly as a function of soil composition, degree of hydration, density and physiological state of host bacteria, and rates of free-phage inactivation. Nevertheless, we predict that the basic principle of phage solid-phase population growth, i.e., a phage-diffusion mediated expanding sphere of bacterial infection, could still apply (13). Thus, without active mixing of soil, e.g., via the action of invertebrates or other localized soil disruptions, we speculate that bacterial microcolonies (or "microsites" using the terminology of 54) within the rhizosphere may display periods of boom or bust with regard to phage attack, with increasing microcolony size, or mere time (i.e., chance), increasing the likelihood of phage-microcolony encounter. Infection of one bacterium within a localized bacterial clone could result in the destruction of part or all of a genetically homogeneous bacterial microcolony. Means by which such coordinated attack may be thwarted could include (i) variation in the physiological or anatomical state of the bacteria making up a clone, so that not all bacteria are equally susceptible to phage attack (including the formation of spores for those species that are able to, as well as hyphael aging for streptomycetes; 13); (ii) display of motility such that bacteria progeny minimize co-location and thereby avoid serial infection (though with the caveat that active movement through soil might increase the likelihood that individual bacteria encounter phage); and (iii) sequestration away from the soil such as within root nodules colonized by rhizobia or perhaps following bacterial penetration into plants upon infection (16). Rarely encountering microcolonies could be antithetical to the prosperity or even survival of obligately lytic phage (54), but could provide numerous stable, otherwise phage-free niches for temperate-phage survival as bacterial lysogens.
The Phage Phyllosphere
How phage interact with their bacterial hosts in the phyllosphere, the aerial plant structures, is even less understood than the phage ecology of the rhizosphere. The phyllosphere presents a less hospitable environment -- relative to the rhizosphere -- given the exposure to UV, intense visible light, and desiccation that is likely on many above-soil plant surfaces (5,60). The harshness of the phyllosphere combined with its relative impermanence begs the question, particularly for obligately lytic phage: Where is the virion reservoir?
In the case of phages of the plant-pathogen Erwinia amylovora (Fig. 4), some studies have noted that phage are less readily isolated from the aerial portions of trees, even during times of active infection by the host (20,24). By contrast, phage could almost always be isolated from the soil around infected trees. That would suggest that the phage reservoir is located in the soil, possibly with phage multiplying on stray bacteria that fall from the tree to the ground below. Other phage studies on the same host bacterium, however, have found abundant E. amylovora phages in the phyllosphere of infected trees (61) (for review of similar observations in additional phage-plant pathogen systems, see: 16). If the virion reservoir is the soil for at least some phage that attack plant epiphytes, then how do phage reach the phyllosphere of a tree, which may have foliage 3 or more feet off the ground? One possible explanation is that phage invade the phyllosphere upon plant germination and then remain a part of plant normal flora. Alternatively, phages could move from plant to plant within the phyllosphere with soil remaining a phyllosphere phage sink rather than source habitat. We simply do not know.
| |

Fig. 4. Erwinia amylovora phage isolate PEa 31-2. Courtesy of Ron Smith and Antonet Svircev. |
|
The habitats of otherwise plant-associated phage can range beyond plants or plant-associated soil. Irrigation waters (Yoshimura & Morihashi, 1959 as cited by 52) and agricultural drainage (66,67) are known to contain phage capable of infecting plant-associated bacteria. Erwinia phage have been isolated from lakes (18) as well as from sewage (76,77). The latter phage perhaps display broad host ranges, attacking enteric bacteria associated with humans as well as the bacteria associated with plants (57). Perhaps similarly, phage infecting phytopathogenic Pseudomonas have been isolated from sewage (43,75) while Agrobacterium-infecting phage have been isolated from feces (52,62).
Erwinia phage have also been isolated from a corn flea beetle (83). This association between phage and insect is of interest since arthropods are known vectors for viruses that directly infect plants. However, the degree to which arthropods are responsible for phage transmission remains an open question (20). Indeed, phage movement within and between plants as well as between soil and plants presumably follows paths similar to those employed by bacteria, i.e., carriage by animals, dust, soil, seeds, and water, including splashing caused by hard rain, plus various human activities such as pruning (26). A case can be made that rain, by promoting epiphytic bacterial growth, can simultaneously supply phage with (i) healthy hosts, (ii) water in which to diffuse between bacteria, and (iii) a means of disseminating about individual plants as well as among populations of plants.
Phage and Agriculture
Though plants are surrounded by phage, the vast bulk of the phage impact on plants is mediated through plant-associated bacteria. Plant-associated symbiotic bacteria can range from helpful (mutuals) to harmful (pathogens), and the phage impact on bacteria also can range from mutualistic to parasitic. Despite these complications, the phage impact, either negative or positive on plants, tends to be limited (i) to phage-induced bacterial lysis, (ii) to selection for phage-resistance within bacterial communities, or (iii) to phage-associated modification of bacterial phenotypes (phage conversion). These we consider in order:
(i) There have been a number of reports of phage presence, in relatively small-scale experiments, that result in reduced plant growth or reduction in plant nitrogen content (e.g., 6,28,32). Experimentally induced decline in the plant-protective bacterium, Pseudomonas fluorescens, has also been noted (37). However, it is uncertain how often wild phages negatively impact on plant growth under naturally occurring conditions. We speculate that extrapolation of observations from small to large scales is challenging due to ignorance of naturally occurring phage-bacteria dynamics and spatial heterogeneities that exist across large plots. Indeed, we are aware of only one report suggesting that phage may have obstructed plant growth on a large scale in a non-experimental agricultural setting, as mediated by reductions in soil rhizobia (Demolon & Dunez, 1935, as cited by 9). Further exploration of this “extrapolation” issue could be difficult assuming reluctance to conduct large-scale field tests of phage that are antagonistic to beneficial bacteria.
(ii) It is typically assumed that reduced plant growth correlated with phage presence is a consequence of phage antagonism against beneficial bacteria, e.g., phage-induced lysis. Lytic phage can also indirectly reduce the fitness of susceptible bacteria. This fitness reduction can be manifest either as a density decline of bacteria inhabiting specific niches, or by a decline only of susceptible bacteria, the latter suggesting a replacement of susceptible bacteria by similar but phage-resistant bacteria. The above-cited Demolon and Dunez (1935) study, for example, has been much discussed in the plant-phage literature, with some authors concluding that the observed negative phage impact resulted from selection for a phage-resistant bacterial phenotype that was less effective at nitrogen fixation (as reviewed by 68). This interpretation is consistent with more modern phage community-ecology theory, which posits that bacterial resistance to phage attack often comes at some metabolic cost to bacteria. Consequently, a bacterium that does not display phage resistance may be able to invade and even drive to extinction a phage-resistant population of otherwise-identical bacteria, at least so long as phage are not present (e.g., 10,39).
From a plant’s perspective, this change in bacterial prevalence upon phage attack is irrelevant unless phage-sensitive and phage-resistant bacteria display differences in their ability to interact with plants. Such differences are often observed, though it is important to note that for many studies only a fraction of bacterial mutations to phage resistance result in significant change in plant-interaction phenotypes. Phage T4- and phage fEC2-resistant, mostly lypopolysaccharide (LPS)-defective mutants of the soft-rot Erwinias E. carotovora and E. chrysanthemi, for example, generally do not display a loss of virulence (38 of 40 and 7 or 9 mutants, respectively) (64,78). Given a relative rarity of avirulence in phage-resistant mutants, an invasion of phage into a pathogen population and subsequent selection for phage-resistant variants may not impact negatively on the virulence of the surviving bacteria. Rather, the community impact may be seen as a decline of the pathogen population followed by recovery by similarly virulent but phage-resistant bacteria.
Conversely, phage-resistant mutants of Ralstonia solanacearum, which display various defects in LPS synthesis, were predominantly avirulent in tobacco seedlings (29), as were approximately 50% of selected phage-resistant Xanthomonas campestris mutants (59). Mutational phage resistance resulting from a loss of pili also has been shown to reduce Rhizobium nodulation in clover (38). Mutation to phage-resistance by P. fluorescens similarly can result in reduced protection of radishes from Fusarium wilt. The mechanism by which this loss occurs, however, appears to be not so much from a decline in P. fluorescens colonizing ability as due to insufficient induction of cross-reactive systemic resistance by plants (45).
(iii) Like phage resistance, modification of bacterial phenotype can result from phage conversion, i.e., the expression of prophage genes over the course of lysogenic infection. Little is known of phage conversion positively affecting plant growth. There exists some evidence for the converse, however: lysogeny negatively impacting soybean-Bradyrhizobium japonicum interaction (1). An example of phage conversion that could possibly be interpreted as positively affecting plant fitness, by poisoning a plant predator, occurs in the case of toxic annual ryegrass, Lolium rigidum. The developing seeds of this grass are susceptible to infection and gall formation by a nematode, Anguina funesta. If these nematodes are carrying certain strains of the bacterium Clavibacter toxicus, then the galls will contain corynetoxin. Related to tunicamycin-like antibiotics, corynetoxin inhibits protein glycosylation and can be fatal to grazing animals (i.e., sheep) that consume the infected grass.
Phages, such as the phage NCPPB 3778, are associated with the ability of C. toxicus to produce corynetoxin (53). It is not clear how the phage causes the production of the toxin, as the complicated glycolipid structure of corynetoxins would require an elaborate metabolic pathway to be expressed from the phage genome. However, associations between phage and bacterial virulence factors are quite common, being responsible for much of the virulence associated with such important human pathogens as Corynebacterium diphtheria, Escherichia coli O157:H7 (Fig. 5), and Vibrio cholerae (79). Note that the likelihood of phage conversion of non-toxogenic C. toxicus strains into toxigenic strains seems to be enhanced via the application various herbicides at the same time as phage exposure (42). Also of interest, phage Xf and Xf-2 infection of X. campestris results in an increase in this bacterium’s virulence towards rice (26,33).
|
 |
|
 |
|
|
Fig. 5. LG1 was isolated from chicken feces on an E. coli O157:H7 host, but it plaques on a wide range of E. coli, and a few Enterobacteriaceae. The black bars are size bars representing 100 nm. The second image shows the same phage with the tail sheath contracted. Image courtesy of Larry Goodridge (25). |
|
Phage Therapy
Phage have been proposed as plant-pathogen control agents in a process known as phage therapy: the application of specific phages to specific ecosystems in order to reduce the population size of specific bacteria. That is, phage therapy is a form of biological control -- the use of one organism to suppress another. Like other methods of biological control, one advantage of phage therapy is a reduction in the usage of chemical agents against pest species, which, in the case of phage, means a reduction in the usage of chemical antibiotics.
Phage therapy was explored extensively by early phage workers as a means of controlling plant pathogens (52). Nevertheless, by 1990 Goto (26) wrote very pessimistically on the subject:
Practical use of phages for control of bacterial plant disease in the field has not been successful. When some control was achieved, this was brought about by inoculation with a mixture of the phage and the bacterium, or by plant or seed treatment with phage before challenge with bacteria. In practice, however, the pathogenic bacteria in plant tissues are in a dense mass and frequently surrounded by abundant extracellular polysaccharides, which prevent effective adsorption of phage particles. Another obstacle is the complexity of phage-bacterium interrelationships in nature, due to the diversity of bacterial strains that differ in phage susceptibility. (p. 91)
Though these problems with phage therapy are not limited to the treatment of plant diseases (71), nonetheless over the past decade the promise of phage therapy as a means of combating plant disease has been explored with increasing enthusiasm.
Circumstances in which phage therapy of plants or plant products has been attempted include against Salmonella associated with fresh-cut fruit (47), to disinfest Streptomyces scabies-infected potato seed-tuber (48), against bacterial leaf spot of mungbeans (particularly in combination with streptomycin; 11), against Xanthomonas pruni-associated bacterial spot of peaches (17,59), to control of X. campestris infections of peach trees (58) as well as cabbage and pepper diseases (74), to control Ralstonia solanacearum (73) (see also 80), and to control soft rot and fire blight associated with Erwinia (19) (see also 20) (Fig. 6). Phage therapy has been used successfully against bacterial blotch of mushrooms caused by Pseudomonas tolaasii (50). In studies notable for the employment of phage host-range mutants, phage therapy has also been employed against bacterial blight of geraniums and bacteria spot of tomatoes, both caused by pathovars of X. campestris (21,22). Phage can also be used to bias the survival of more-effective mutualistic bacteria. Basit et al. (9), for example, have isolated phage that are ineffective against a preferred inoculum of B. japonicum but effective against naturally occurring competitors. By coating seeds with phage effective only against these potential competitors they can enhance nitrogen fixation (see also 51).
| |

Fig. 6. Escherichia coli under mass attack by numerous phage-T4 virions. |
|
Though seemingly effective in certain situations, it is likely that phage therapy against bacterial plant pathogens will not prove to be a “magic bullet” in all cases. Johnson (32) proposed a general biological control model which suggests that the success of a particular treatment will be influenced by agent and target densities. An important component of this model is the possibility of the target residing in spatial refuges into which the biological control agent cannot penetrate. We would propose several additional factors that could contribute to the success or failure of a potential phage therapy system, such as the location or niche in which the target pathogen population resides (including the potential for refuges), the presence of adequate water as a medium for virion diffusion, rates of virion decay, the timing of phage application (55,56), phage in situ infection fecundity, and the relative fitness (and virulence) of phage-resistant bacterial mutants. Furthermore, due to the diversity of bacteria and their phages, extrapolation of phage therapy practices from one pathogen system to different systems will not always be practicable. Indeed, our nominal understanding of various aspects of the phage ecology of plants could hinder the development of phage therapy regimens that function reliably under diverse circumstances.
Acknowledgment
Thanks to Bruce Abedon for reading and commenting on the manuscript.
Additional Resources
The Bacteriophage Ecology Group
The Bacteriophage Home Page
Bacteriophage Names 2000-A list of phage names and discussion of phage naming
The International Committee on Taxonomy of Viruses site
The Big Picture Book of Viruses
Selected References
1. Abebe, H. M., Sadowsky, M. J., Kinkle, B. K., and Schmidt, E. L. 1992. Lysogeny in Bradyrhizobium japonicum and its effect on soybean nodulation. Appl. Environ. Microbiol. 58:3360-3366.
2. Abedon, S. T. 1990. Selection for lysis inhibition in bacteriophage. J. Theor. Biol. 146:501-511.
3. Abedon, S. T. 2004. Phage ecology. In: The Bacteriophages. R. Calendar, ed. Oxford University Press, Oxford. In press.
4. Ackermann, H. W. 1997. Bacteriophage ecology. Pages 335-339 in: Progress in Microbial Ecology (Proceedings of Seventh International Symposium on Microbial Ecology). M. T. Martins, M. I. Z. Sato, J. M. Tiedje, L. C. N. Hagler, J. Döbereiner, and P. S. Sanchez, eds. Brazilian Society for Microbiology.
5. Adams, M. H. 1959. Bacteriophages. Interscience, New York.
6. Ahamd, M. H., and Morgan, V. 1994. Characterization of a cowpea (Vigna unguiculata) rhizobiophage and its effects on cowpea nodulation and growth. Biol. Fertil. Soils 18:297-301.
7. Arber, W. 1983. A beginner's guide to lambda biology. Pages 381-394 in: Lambda II. R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg, eds. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
8. Ashelford, K. E., Day, M. J., and Fry, J. C. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289.
9. Basit, H. A., Angle, J. S., Salem, S., and Gewaily, E. M. 1992. Phage coating of soybean seed reduces nodulation by indigenous soil bradyrhizobia. Can. J. Microbiol. 38:1264-1269.
10. Bohannan, B. J. M., and Lenski, R. E. 2000. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecology Letters 3:362-377.
11. Borah, P. K., Jindal, J. K., and Verma, J. P. 2000. Integrated management of bacterial leaf spot of mungbean with bacteriophages of Xav and chemicals. J. Mycol. Plant Pathol. 30:19-21.
12. Burge, W. D., and Enkiri, N. K. 1978. Virus adsorption by five soils. J. Environ. Qual. 7:73-76.
13. Burroughs, N. J., Marsh, P., and Wellington, E. M. H. 2000. Mathematical analysis of growth and interaction dynamics of streptomycetes and a bacteriophage in soil. Appl. Environ. Microbiol. 66:3868-3877.
14. Calendar, R. 2004. The Bacteriophages. Oxford University Press, Oxford.
15. Chattopadhyay, D., and Puls, R. W. 2000. Forces dictating colloidal interactions between viruses and soil. Chemosphere 41:1279-1286.
16. Civerolo, E. L. 1972. Interaction between bacteria and bacteriophages on plant surfaces and in plant tissues. Pages 25-37: Plant Pathogenic Bacteria 1971. The Netherlands.
17. Civerolo, E. L., and Kiel, H. L. 1969. Inhibition of bacterial spot of peach foliage by Xanthomonas pruni bacteriophage. Phytopathology 59:1966-1967.
18. Eayre, C. G., Bartz, J. A., and Concelmo, D. E. 1995. Bacteriophages of Erwinia carotovora and Erwinia ananas isolated from freshwater lakes. Plant Dis. 79:801-804.
19. Eayre, C. G., Concelmo, D. E., and Bartz, J. A. Control of soft rot Erwinias with bacteriophages. Phytopathology 80[10], 994. 1990. (Abstract)
20. Erskine, J. M. 1973. Characteristics of Erwinia amylovora bacteriophage and its possible role in the epidemiology of fire blight. Can. J. Microbiol. 19:837-845.
21. Flaherty, J. E., Harbaugh, B. K., Jones, J. B., Somodi, G. C., and Jackson, L. E. 2001. H-mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. Hortscience 36:98-100.
22. Flaherty, J. E., Jones, J. B., Harbaugh, B. K., Somodi, G. C., and Jackson, L. E. 2000. Control of bacterial spot on tomato in the greenhouse and field with h-mutant bacteriophages. Hortscience 35:882-884.
23. Friedman, D. I., and Gottesman, M. 1983. Lytic mode of lambda development. Pages 21-51 in: Lambda II. R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg, eds. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
24. Gill, J. J., Svircev, A. M., Smith, R., and Castle, A. J. 2003. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 69:2133-2138.
25. Goodridge, L., Gallaccio, A., Griffiths, M.W. (2003). Morphological, host range, and genetic characterization of two coliphages. Applied and Environmental Microbiology 69(9):5364-5371.
26. Goto, M. 1992. Fundamentals of Bacterial Plant Physiology. Academic Press, New York.
27. Gvozdyak, R. I. 1993. Plant, phage, bacterium: A new hypothesis on their interrelation. Mikrobiologicheskii Zhurnal (Kiev) 55:92-94.
28. Hammad, A. M. M. 1998. Evaluation of alginate-encapsulated Azotobacter chroococcum as a phage-resistant and an effective inoculum. J. Basic Microbiol. 38:9-16.
29. Hendrick, C. A. and Sequeira, L. 1984. Lipopolysaccharide defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94-101.
30. Henning, U. and Hashemolhosseini, S. 1994. Receptor recognition by T-even type coliphages. Pages 291-298 in: The Molecular Biology of Bacteriophage T4. J. D. Karam, ed. ASM Press, Washington, DC.
31. Hussein, M. E., El-Hawa, M. E. A., and El Dydamony, G. 1994. Population and persistence of Zag-1 phage and cowpea Rhizobium in two sterile soils. Egyptian Journal of Microbiology 29:271-283.
32. Johnson, K. B. 1994. Dose-response relationships and inundative biological control. Phytopathology 84:780-784.
33. Kamiunten, H. and Wakimoto, S. 1983. Effects of infection with filamentous phage Xf on the growth, ultrastructure, and virulence of Xanthomonas capestris var. oryzae N-5850. An. Phytopathol. Soc. Japan 48:642-647.
34. Kaplan, D. A., Naumovski, L., Rothschild, B., and Collier, R. J. 1981. Appendix: A model of plaque formation. Gene 13:221-225.
35. Karam, J. D. 1994. Molecular Biology of Bacteriophage T4. ASM Press, Washington, DC.
36. Kasman, L. M., Kasman, A., Westwater, C., Dolan, J., Schmidt, M. G., and Norris J. S. 2002. Overcoming the phage replication threshold: A mathematical model with implications for phage therapy. J. Virol. 76:5557-5564.
37. Keel, C., Ucurum, Z., Michaux, P., Adrian, M., and Haas, D. 2002. Deleterious impact of a virulent bacteriophage on survival and biocontrol activity of Pseudomonas fluorescens strain CHAO in natural soil. Mol. Plant-Microbe Interact. 15:567-576.
38. Kleczkowska, J. 1950. A study of phage resistant mutants of Rhizobium trifolii. J. Gen. Microbiol. 4:298-310.
39. Kleczkowska, J. 1957. A study of the distribution and the effects of bacteriophage of root nodule bacteria in the soil. Can. J. Microbiol. 3:171-180.
40. Koch, A. L. 1964. The growth of viral plaques during the enlargement phase. J. Theor. Biol. 6:413-431.
41. Krylov, V. 2003. The role of horizontal gene transfer by bacteriophages in the origin of pathogenic bacteria. Russian Journal of Genetics 39:483-504.
42. Kurtböke, D. I. 1994. Rapid detection of Clavibacter toxicus and of its bacteriophage responsible for annual ryegrass toxicity in Australia and the effect of selected herbicides on toxin production. Actinomycetes 5:31-39.
43. Lee, I. F., and Boezi, J. A. 1966. Characterisation of bacteriophage gh-1 for Pseudomonas putida. J.Bacteriol. 92:1821-1827.
44. Lee, Y., Eisner, S. D., and Yin, J. 1997. Antiserum inhibition of propagating viruses. Biotech. Bioeng. 55:542-546.
45. Leeman, M., Van Pelt, J. A., Den Ouden, F. M., Heinsbroek, M., Bakker, P. A. H., and Schippers, B. 1995. Induction of systemic resistance against fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85:1021-1027.
46. Lepault J., Leonard K. (1985) Three-dimensional structure of unstained, frozen-hydrated extended tails of bacteriophage T4. Journal of Molecular Biology 182(3):431-41.
47. Leverentz, B., Conway, W. S., Alavidze, Z., Janisiewicz, W. J., Fuchs, Y., Camp, M. J., Chighladze, E., and Sulakvelidze, A. 2001. Examination of bacteriophage as a biocontrol method for salmonella on fresh-cut fruit: A model study. J.Food Prot. 64:1116-1121.
48. McKenna, F., El-Tarabily, K. A., Hardy, G. E. S. T., and Dell, B. 2001. Novel in vivo use of a polyvalent Streptomyces phage to disinfest Streptomyces scabies-infected seed potatoes. Plant Pathol. 50:666-675.
49. Menzel, G., Stenz, E., Toure, I. M., Gebler, B., and Schuster, G. 1975. Effect of several plant growth regulators on various prokaryotes and their viruses. Zeitschrift fur Allgemeine Mikrobiologie 15:259-268.
50. Munsch, P., and Olivier, J. M. 1995. Biocontrol of bacterial blotch of the cultivated mushroom with lytic phages: Some practical considerations. Pages 595-602 in: Science and Cultivation of Edible Fungi, Vol. II: Proceedings of the 14th International Congress. T. J. Elliott, ed.
51. Novikova, N. I., and Simarov, V. B. 1990. Competitiveness of alfalfa Rhizobia in the presence of a bacteriophage. Doklady Vsesoyuznoi Ordena Lenina i Ordena Trudovogo Krasnogo Znameni Akademii Selskokhozyaistvennykh Nauk Imeni V I Lenina 29-31.
52. Okabe, N., and Goto, M. 1963. Bacteriophages of plant pathogens. Ann. Rev. Phytopathol. 1:397-418.
53. Ophel, K. M., Bird, A. F., and Kerr, A. 1993. Association of bacteriophage particles with toxin production by Clavibacter toxicus, the causal agent of annual ryegrass toxicity. Phytopathology 83:676-681.
54. Pantastica-Caldas, M., Duncan, K. E., Istock, C. A., and Bell, J. A. 1992. Population dynamics of bacteriophage and Bacillus subtilis in soil. Ecology 73:1888-1902.
55. Payne, R. J. H., and Jansen, V. A. A. 2003. Pharmacokinetic principles of bacteriophage therapy. Clinical Pharmacokinetics 42:315-325.
56. Payne, R. J. H., Phil, D., and Jansen, V. A. A. 2000. Phage therapy: The peculiar kinetics of self-replicating pharmaceuticals. Clinical Pharmacology and Therapeutics 68:225-230.
57. Pirhonen, M., and Palva, E. T. 1988. Occurence of bacteriophage T4 receptor in Erwinia carotovora. Mol. Gen. Genet. 214:170-172.
58. Randhawa, P. S., and Civerolo, E. L. Inhibition of Xanthomonas campestris pathovar pruni by bacteria and pruniphage on detached peach leaves. Phytopathology 74:864. 1984. Abstract
59. Randhawa, P. S. and Civerolo, E. L. 1986. Interaction of Xanthomonas campestris pv pruni with pruniphages and epiphytic bacteria on detached peach trees. Phytopathology 76:549-553.
60. Ritchie, D. F. 1978. Bacteriophages of Erwinia amylovora: Their Isolation, Distribution, Characterization, and Possible Involvement in the Etiology and Epidemiology of Fire Blight. Michigan State University.
61. Ritchie, D. F., and Klos, E. J. 1977. Isolation of Erwinia amylovora bacteriophage from aerial parts of apple trees. Phytopathology 67:101-104.
62. Roslycky, E. B. 1962. Phages for Agrobacterium radiobacter with reference to host range. Can. J. Microbiol. 8:71-78.
63. Sato, M. 1983. Phage induction from lysogenic strains of Pseudomonas syringae pathovar mori by the extract from mulberry leaves. An. Phytopathol. Soc. Japan 49:259-261.
64. Schoonejans, E., Expert, D., and Toussaint, A. 1987. Characterisation and virulence properties of Erwinia chrysanthemi lipopolysaccharide-defective fEC2-resistant mutants. J. Bacteriol. 169:4011-4017.
65. Schrag, S., and Mittler, J. E. 1996. Host-parasite persistence: the role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 148:348-347.
66. Smit, E., van Elsas, J. D., Van Veen, J. A., and de Vos, W. M. 1991. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil by using bacteriophage fR2f for donor counterselection. Appl. Environ. Microbiol. 57:3482-3488.
67. Smit, E., Wolters, A. C., Lee, H., Trevors, J. T., and van Elsas, J. D. 1996. Interactions between a genetically marked Pseudomonas fluorescens strain and bacteriophage PHI-R2f in soil: Effects of nutrients, alginate encapsulation, and the wheat rhizosphere. Microb. Ecol. 31:125-140.
68. Staniewski, R., Kowalski, M., Gogaoz, E., and Sokolowska, F. 1962. Susceptibility of Rhizobium strains to phages. Acta Microbiologica Polonica 11:245-254.
69. Stewart, F. M., and Levin, B. R. 1984. The population biology of bacterial viruses: Why be temperate. Theor. Pop. Biol. 26:93-117.
70. Storey, M. V., and Ashbolt, N. J. 2001. Persistence of two model enteric viruses (B40-8 and MS-2 bacteriophages) in water distribution pipe biofilms. Water Sci. Technol. 43:133-138.
71. Summers, W. C. 2001. Bacteriophage therapy. Ann. Rev. Microbiol. 55:437-451.
72. Sykes, I. K., Lanning, S., and Williams, S. T. 1981. The effect of pH on soil actinophage. J. Gen. Microbiol. 122:271-280.
73. Tanaka, H., Negishi, H., and Maeda, H. 1990. Control of tobacco bacterial wilt by an avirulent strain of Pseuomonas solanacearum M4S and its bacteriophage. An. Phytopathol. Soc. Japan 56:243-246.
74. Tanaka, H., Negishi, H., and Maeda, H. 1990. Control of tobacco bacterial wilt by an avirulent strain of Pseuomonas solanacearum M4S and its bacteriophage. An. Phytopathol. Soc. Japan 56:243-246.
75. Thomas, M. D., and Leary, J. V. 1983. Bacteriophages from sewage specific for fluorescent phytopathogenic pseudomonads. Phytopathology 73:403-406.
76. Toth, I. K., Hyman, L. J., Pérombelon, M. C. M., and Salmond, G. P. C. 1989. A generalized transduction system for Erwinia carotovora and the use of phages to isolate reduced virulence (Rvi-) mutants on potato. Pages 801-806 in: Proceedings of the International Conference on Plant Pathogenic Bacteria 7th. Z. Klement, ed. Academiai Kiado, Budapest, Hungary.
77. Toth, I. K., Mulholland, V., Cooper, V., Bentley, S., Shih, Y. L., Perombelon, M. C. M., and Salmond, G. P. C. 1997. Generalized transduction in the potato blackleg pathogen Erwinia carotovora subsp. atroseptica by bacteriophage fM1. Microbiology (Reading) 143:2433-2438.
78. Toth, I. K., Thorpe, C. J., Bentley, S. D., Mulholland, V., Hyman, L. J., Perombelon, M. C. M., and Salmond, G. P. C. 1999. Mutation in a gene required for lipopolysaccharide and enterobacterial common antigen biosynthesis affects virulence in the plant pathogen Erwinia carotovora subsp. atroseptica. Mol. Plant-Microbe Interact. 12:499-507.
79. Wagner, P. L., and Waldor, M. K. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993.
80. Wall, G. C., Sanchez, J. L., Hartman, G. L., and Hayward, A. C. 1993. A biocontrol agent for Pseudomonas solanacearum. ACIAR Proceedings 320-321.
81. Williams, S. T., Mortimer, A. M., and Manchester, L. 1987. Ecology of soil bacteriophages. Pages 157-179 in: S. M. Goyal, C. P. Gerba, and G. Bitton, eds. Phage Ecology. John Wiley & Sons, New York.
82. Wommack, K. E. and Colwell, R. R. 2000. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114.
83. Woods, T. L., Israel, H. W., and Sherf, A. F. 1982. Isolation and partial characterization of a bacteriophage of Erwinia stewartii from the corn flea beetle Chaetocnema-pulicaria. Protec. Ecol. 3:229-236.
84. Yin, J., Duca, K., Lam, V., Keren, I., Endler, E. E., Letchworth, G. J., and Novella, I. S. 2001. Quantifying viral propagation in vitro: toward a method for characterization of complex phenotypes. Biotechnol. Prog. 17:1156-1165.
85. Yin, J., and McCaskill, J. S. 1992. Replication of viruses in a growing plaque: A reaction-diffusion model. Biophys. J. 61:1540-1549.
86. You, L., and Yin, J. 1999. Amplification and spread of viruses in a growing plaque. J. Theor. Biol. 200:365-373.
