|
|

Figure 1.
Caenorhabditis
elegans adult and two juveniles.
Click image for a larger view. |
A milestone in an animal
Recently, the free-living nematode
Caenorhabditis elegans (FIGURES 1,3-6) became the first animal and more importantly, the first multicellular organism, to have the sequencing of its genome essentially completed (
C. elegans Consortium, Science 282:2011-2045, 1998). This is a landmark accomplishment for all of biology since we can now begin to investigate the phenomena that made cells come together and function in a complex multicellular system. The genetic blueprint (DNA) of
C. elegans consists of ~97 million base pairs mapped onto six pairs of chromosomes and encodes >19,099 proteins contained in a mere 959 cells (among which are 302 neurons). This provides biologists for the first time with a view of all the genes present in an animal. The only previous eukaryote with a sequenced genome is the yeast
Saccharomyces cerevisiae, which is unicellular (Goffeau et al. 1996). Proteins unique to the nematode (and not yeast) may well define metazoans (Chervitz et al., 1998). Other comparisons of bacterial, yeast, nematode, plant, mouse and human genomes will reveal unique and surprising aspects of the genetic make-up of organisms.
 |
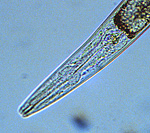 |
Figure 2. Anterior of plant parasitic nematode
Meloidogyne incognita juvenile
showing protrusible spear (stylet).
Click image for a larger view |
Figure 3. Anterior of
C. elegans showing
cylindrical mouth cavity (for feeding on bacteria)
Note stylet is absent.
Click image for a larger view |
Since Sydney Brenner proposed
C. elegans as a model metazoan for study of higher organisms in 1965, a wealth of knowledge has been generated in genetics, neurobiology, cell and molecular biology, genomics and bioinformatics, aging, cell death, signal transduction, muscle contraction, behavior, digestion, reproduction and development (Epstein and Shakes, 1995; Riddle et al., 1997; Wood and the Community of
C. elegans researchers, 1988). However, any recognition of the pioneers of the genomic era will include the efforts of Coulson, Sulston, and Waterston (among others such as Venter). Suslton and Coulson provided much of the fundamental biological and genetic data to interpret the biology of the emerging genome (Pennisi, 1998).
Nematodes are non-segmented roundworms and comprise the most abundant metazoan (animal) life form (Baldwin et al., 1997; Lambshead, 1993). Parasitic nematodes threaten the health of plants, animals and humans worldwide, but the majority of nematode species play important beneficial roles, for example, as secondary consumers in soil and aquatic ecosystems (Niles and Freckman, 1998). Estimates of crop losses caused by plant-parasitic nematodes exceed $77 billion worldwide and $8 billion (or ~12%) in the USA (Sasser and Freckman, 1987). Plant-parasitic nematodes are obligate parasites that cannot be grown on plates of bacteria as a food source to provide large numbers of individuals as can certain free-living nematodes (e.g.
C. elegans ). Phylogenetic analysis using small subunit ribosomal DNA sequences from diverse nematode species suggests that plant parasitism evolved independently at least three times in phylum Nematoda, viz. Orders Dorylaimida and Triplonchida in Class Adenophorea and Order Tylenchida (Blaxter et al., 1998) and possibly Order Aphelenchida in Class Secernentea. The most conspicuous evolutionary adaptations of nematodes for plant parasitism include the development of a protrusible feeding spear (called a stylet) (FIGURE 2) and major morphological and physiological modifications of the esophagus (Hussey and Williamson, 1998).
The transparent body of
C. elegans, its near-microscopic size (<1 mm), ease of culture and rapid life cycle simplified questions raised in the study of systems in humans, mice and even fruit flies. The nematode produces adult hermaphrodites that allow both outcrossing and selfing for genetic analyses. The developmental fate and connections of each of the nearly 1,000 cells in the adult nematode are known. Although more than half of the newfound
C. elegans genes have no identified function, researchers plan to inactivate every gene to understand its role in the nematode.
 |
 |
Figure 4. Developing embryos in the uterus of the nematode C. elegans. Click image for a larger view |
Figure 5.Caenorhabditis elegans embryo at 1.5-fold stage inside egg. Click image for a larger view |
 |
Figure 6. Embryonic development of the nematode
C. elegans within the eggshell.
Photograph courtesy Fabio Piano. Click image for a larger view and more information. |
Implications for plant nematology
Although many of the genetic techniques used by
C. elegans researchers may not be immediately applicable to plant-parasitic nematodes, there are several important areas in which the accomplishments of the
C. elegans genome project can affect plant nematology: gene cloning, transformation strategies, gene homologs, gut antigens, dauer larva biology, synteny cloning, genome sequencing, evolution of plant parasitism, the nervous system and host finding, to name a few.
Gene cloning
The availability of the
C. elegans genome sequence will facilitate isolation of genes of interest in plant-parasitic nematodes by using genes cloned from
C. elegans as probes. The isolation of genes controlling nematode surface identity is one example demonstrating the utility of
C. elegans genetic information. Collagens and cuticulins are important structural proteins in nematode cuticles (Koltai et al., 1997; DeGiorgi et al., 1997; Ray et al., 1996). During molting and development, the cuticle of plant-parasitic nematodes undergoes biochemical changes. A probe made from a
C. elegans cuticulin gene (
Cut-1) was used to screen a genomic library of the parasitic root-knot nematode
Meloidogyne artiellia. Sequence analysis revealed very similar promoter regions, and 75% homology at the amino acid level (DeLuca et al., 1994). The promoter regions of collagen genes (
Col-2 and
Col-6) were also highly homologous between
C. elegans and
M. artiellia. For less conserved gene sequences, PCR-based approaches can be designed using degenerate primers. Primers may also be designed on the basis of partial amino acid sequences of a gene product. The resultant PCR product can be used as a probe to isolate the gene of interest (Jones et al., 1997).
Transformation
Efficient transformation systems for plant-parasitic nematodes need to be developed to study the regulation and localization of expressed genes. DNA transformation (involving microinjection of DNA into adult gonads) has been accomplished for
C. elegans (Mello and Fire, 1995). Several animal-parasitic genes have been introduced and expressed in
C. elegans (Grant, 1992; Riddle and Georgi, 1990). A GAPDH promoter from the potato cyst nematode was expressed in
C. elegans (Qin et al., 1998). Basic molecular and developmental mechanisms may have been conserved across nematode genera and so transformation of
C. elegans with "parasitism" genes (such as those associated with esophageal gland secretions) may elucidate mechanisms of transcriptional regulation in plant-parasitic nematodes (Hussey et al., 1994). Alternatively, the absence of a nematode parasitism gene from
C. elegans, like the cellulases of cyst nematodes, may imply a different origin of some parasitism genes (Davis et al., 2000). The degree of conservation between animal genomes is so high that it is often possible to find homologs of
C. elegans genes in mice or humans (The
C. elegans sequencing consortium, 1998).
Gut antigens
Gut surface antigenic epitopes may be highly conserved among nematode genera and could provide protective immunity in the case of animal parasites (Jasmer et al. 1993; Smith et al. 1993). Gut receptors of plant-parasitic nematodes that are important in feeding may have their closest homologs in free-living soil nematodes like
C. elegans, cephalobes or herbivorous insects. Gut epitopes and corresponding genes are potentially useful targets for inactivation and engineering resistance to plant-parasitic nematodes.
Dauer larva development
One pathway in
C. elegans that will be useful for study by plant nematologists is the development of the dauer larval stage (Blaxter and Bird, 1997; Riddle and Albert, 1997). The dauer (enduring) stage of
C. elegans is induced when unfavorable environmental conditions exist (e.g., limited food supply). The dauer is developmentally arrested and specialized for long-term survival and dispersal (Riddle and Albert, 1997). Genes controlling
dauer
formation (
daf genes) have been extensively analyzed in
C. elegans (Riddle and Albert, 1997). Certain juvenile stages of plant-parasitic nematodes exhibit dauer-like characteristics (example root-knot, cyst, and pine wood nematodes), suggesting that the dauer pathway may be a fundamental aspect of nematode biology (Bird and Opperman, 1998). The signal to exit the dauer stage prior to feeding by
C. elegans is presumed to be similar to the one for root-knot and cyst nematodes (Bird and Opperman, 1998). Nematode developmental decisions are based on environmental cues. Thus, the availability of the sequence of the whole
C. elegans genome and the likely existence of homologs of
daf genes in plant-parasitic nematodes, make studies on the dauer pathway in plant-parasitic nematodes worthwhile.
ASPs (Ancylostoma-secreted proteins like ASP-1 and ASP-2) are associated with the switch from dauer state to parasitism when the hookworm
Ancylostoma caninum enters a host. It is envisioned that genes encoding ASPs are generally conserved in parasitic nematodes and could have homologs or orthologs in
C. elegans (Bird and Opperman, 1998). The gene
asp-1 has a homolog in
C. elegans, however, preliminary analyses indicate that there is no obvious homolog of
asp-2 in
C. elegans suggesting
asp-2 has essential parasitic function (Bird and Opperman, 1998). An
asp-2 homolog does exist in the plant parasite
Meloidogyne incognita (Ding et al., 2000).
Synteny cloning
Despite the preeminence of
C. elegans as a model nematode, it is not the best model of a plant parasite. The strongest efforts to understand the genetics of parasitism have centered on cyst nematodes (
Globodera rostochiensis and
Heterodera glycines) (Rouppe van der Voort et al., 1994; Dong and Opperman, 1997). A clever approach to circumvent problems with gene-cloning and large divergence is the exploitation of conserved synteny, the colinearity of homologous loci between nematode species (Blaxter, 1998; Bird and Opperman, 1998). Rhabditids (which include
C. elegans and cephalobes) and tylenchids (which comprise the vast majority of plant-parasitic nematodes) are sister taxa (Blaxter et al., 1998; Blaxter, 1998). The mapping of synteny will enable predictions about gene positions in plant parasites and cephalobes based on locations of homologous genes in
C. elegans (Bird and Opperman, 1998). Sequencing regions around conserved genes (assuming conserved synteny) should unveil genes of interest that show low conservation.
Genome sequencing effort - a precedent
The
C. elegans genome effort generated new technology and techniques that have been directly applicable to the sequencing and analysis of other animal and even human genomes. The successful
C. elegans system has served as a guide in the development of the soybean cyst nematode as a model plant
parasite and energized efforts in other genome projects such as the Filarial Nematode Genome Project for the parasite
Brugia malayi which causes elephantiasis in humans (Bird et al., 1999). Future sequencing of genomes of other nematodes (such as the cephalobes), which are phylogenetically closer to the plant parasites than to
C. elegans, will be facilitated by the achievements and breakthroughs of the
C. elegans genome
consortium. Although many parasitism genes may not have obvious homologs in
C. elegans, transitive or remnant sequences may be identified in the
C. elegans genome if clear homologs of those genes can be found in other bacterial feeders (such as the cephalobes).
Evolution of plant parasitism
Plant nematology will benefit from immediate investigation of the evolution of plant parasitism in the nematode using the
C. elegans sequence as a starting framework to analyze or predict pathways of transcriptional regulation of orthologs (genes of different species with functional similarity that can be traced to a common ancestor). Most of the core biological functions (RNA and DNA metabolism, protein folding, trafficking, degradation, and intermediary metabolism) of the eukaryotes
C. elegans and yeast
S. cerevisiae appear to be orchestrated by orthologous proteins (Chervitz et al., 1998). As more sequence information is obtained for plant-parasitic nematodes, the main interests of plant nematologists will shift from shared core functions within
C. elegans to functions characteristic of plant parasitism. Although entire proteins may not be conserved, high domain conservation may exist and so identification of both shared domains and unique domain combinations should provide insights into functional divergence between
C. elegans (free-living) and plant-parasitic forms. Since the evolutionary divergence between yeast,
C. elegans and humans does not prevent or interfere with finding of orthologs and shared domains (Chervitz et al., 1998), it is envisaged that functional annotations for a future plant-parasitic nematode DNA sequence will be very reliable. A recent hypothesis for the evolution of plant parasitism suggest that, since parasitism genes in plant-parasitic nematodes (such as cellulases) are similar to microbial genes, plant parasitism genes were acquired through horizontal gene transfer (Davis et al., 2000). Acquisition of such genes could have been through incorporation of endosymbiont nonphotosynthetic bacteria or protista into progenitor bacterial-feeding nematodes (such as the cephalobes). There is molecular phylogenetic evidence suggesting plant-parasitic nematodes are closer to cephalobes than to bacterial-feeders such
C. elegans (Blaxter et al., 1998).
Bioengineers of crop resistance to plant-parasitic nematodes may need to target genes/gene products that are induced during pathogenesis (viz. genes that affect nervous system function during food source localization, genes involved in the "turning off" of the dauer larval mode, gut receptors involved in food uptake from the alimentary system, and nematode genes involved in feeding site induction). Proteomics (the analysis of the protein complement expressed by a tissue) may facilitate identification of gene products induced in the host plant and nematode during pathogenesis.
Nervous system and host finding
The well-described nervous system of
C. elegans provides opportunities to examine behaviors and isolate/analyze chemo-, thigmo- and thermo-sensory homologs of genes in important plant-parasitic nematodes. The mechanism(s) by which plant-parasitic nematodes find their hosts and feeding sites are not known. The molecules involved in sensory recognition and signaling could be investigated by analyzing chemotaxis-defective mutants and genetic analysis of candidate receptor genes (Bargmann and Mori, 1997).
Since the nervous system contains about one-third of all somatic cells in
C. elegans (Bargmann, 1998), it is probably the most important equipment used by plant-parasitic nematodes in host finding and localization of feeding sites. Interference with host finding by modifying chemotactic responses has been suggested as a potential strategy for protecting crops against plant-parasitic nematodes (Zuckerman and Jansson, 1984). Identification of molecular genetic differences between the nervous system of
C. elegans and a plant-parasitic nematode will facilitate intelligent bioengineering of crop plants that minimizes interference with vertebrate systems. In
C. elegans, ion channels are similar to vertebrate channels, but voltage-activated sodium channels are apparently absent (Bargmann, 1998). Olfactory chemoreceptors, however, are unrelated in sequence (although similar in properties) to those in vertebrates. Isolation of homologs of these olfactory receptors in plant-parasitic nematodes may indicate whether it is feasible to interfere with nematode host finding as a strategy for crop protection. Nematode-specific neuropeptide transmitters, including the widespread class with FMRF-amide (Phe-Met-Arg-Phe-amide) or similar sequence at the COOH-terminus, have been identified in
C. elegans (Cowden, et al., 1993). Mutant
flp-1 (an FMRF-amide gene) caused behavioral defects, including uncoordination, hyperactivity, and insensitivity to high osmolarity in
C. elegans (Nelson et al., 1998).
Distinctions unveiled
About 58% of the putative coding regions of the
C. elegans genome appear to be nematode-specific. This represents ~400 distinct protein domains (Blaxter, 1998). Nematodes differ from other organisms in the following distinct ways (Blaxter, 1998):
(i) About 80% of
C. elegans genes are trans-spliced to a common spliced leader exon.
(ii) About 20% of
C. elegans genes are organized as operons (i.e., cotranscribed sets of two or more genes).
(iii) Nematodes have a functional glyoxylate cycle (that enables formation of carbohydrates from fatty acids) and can synthesize polyunsaturated fatty acids de novo.
(iv) Differences exist in the biosynthesis of the cuticle, for example the existence of SXC (six-cysteine) domains in the surface coat of animal-parasitic nematodes. The SXC motif is most likely involved in protein-protein interactions.
(v) Nematodes possess surface-located lipid-binding proteins (thought to play roles in nutrient scavenging from the host or transport of lipid within animal-parasitic nematodes). Examples include the Nematode polyprotein allergen (NPA) and the Lipid-binding protein (LBP-20) which also has homologs in the plant-parasitic nematode
Globodera pallida.
Outlook
The
C. elegans genome sequence will allow molecular plant nematologists to enumerate presence/absence of plant parasitism-related gene homologs that are broadly present in plant-parasitic groups. At the outset, it will speed up genetic investigations. Simple PCR amplifications using primers designed from the genome sequence will facilitate molecular cloning of genetic regions of interest. Transformation of particular genetic regions in mutants or wild type will reveal any enhancement or suppression of phenotypes. Fusion of DNA sequences (with or without promoter regions) to the green fluorescent protein (GFP) reporter gene has facilitated studies of spatial and temporal expression profiles of
C. elegans genes and screening of mutants (Mello and Fire, 1995; Riddle et al., 1997). It may be possible to prove functional analogy by demonstrating rescue of a mutation in
C. elegans after transformation with a cloned plant-parasitic gene (Bird and Opperman, 1998). Two powerful technologies that can prove the necessity of a gene or its orthology include (i) deliberate construction of hybrid genes to cause misexpression based on deletions in specific genes (Jansen et al., 1997), and (ii) RNA interference (RNAi) wherein candidate genes are inactivated by injection of double stranded RNA (Fire et al., 1998; Guo and Kemphues, 1995).
Future reverse genetics to study parasitism genes in plant-parasitic nematodes will depend on the availability of efficient promoters. Already, PCR-based cloning efforts have resulted in the isolation of at least one potentially valuable promoter from cyst nematodes (Qin et al., 1998). The promoter successfully drove green fluorescent protein report gene expression in
C. elegans.
Synteny cloning of nematode genes may be feasible given the highly conserved gene order observed in
C. elegans and
C. briggsae (Kuwabara and Shah, 1994). Comparison of conserved segments of upstream regions of certain genes could identify promoter activity (Gilleard et al., 1997). Comparative studies on a 65-kb segment surrounding a gene homolog of interest revealed high conservation in local gene order and synteny between
C. elegans and the distantly related filarial parasitic
Brugia (Blaxter, 1998). Thus comparative sequencing around genomic regions of interest will reveal more information on nematode gene evolution and function (Blaxter, 1998) including homologs related to putative parasitism genes. With the significant accomplishments of the
C. elegans community of researchers and technologies developed in other genome projects (such as the Human Genome Project), the future bodes well for research on plant-parasitic nematodes and other organisms. These accomplishments mark only the beginning of the genomics/proteomics era.
Resources celebrating C. elegans
Bird et al. (1999) present a more detailed overview of the C. elegans sequencing project. The following web sites provide easy-to-reach information on the nematode C. elegans, the C. elegans sequencing project and the status of research, and links to other nematode resources:
Genome Sequencing Center, Washington University (USA)
http://genome.wustl.edu
Sanger Center, Cambridge (England)
http://www.sanger.ac.uk/Projects/C_elegans/
Research on evolution of parasitism and molecular phylogenetics
http://nematode.unl.edu
Early development of
Caenorhabditis elegans:
http://www.mbg.cornell.edu/cals/mbg/faculty-staff/faculty/kemphues.cfm
Society of Nematologists
http://www.nematologists.org/
Selected References
1. Baldwin, J.G., Nadler, S.A., and Wall, D.H. 1997. Nematodes: Pervading the Earth and Linking all Life. Pp. 176-191. In: Raven, P.H. (ed.). National Academy Press, Washington, D.C. 625 pp.
2. Bargmann, C. I. 1998. Neurobiology of the
Caenorhabditis elegans genome. Science 282:2028-2033.
3. Bargmann, C. I. And Mori, I. 1997. Chemotaxis and Thermotaxis. Pp. 717-737. In: Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (eds).
C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY 1222 pp.
4. Bird, D.M. and Opperman, C. H. 1998.
Caenorhabditis elegans. J. Nematol. 30:299-308.
5. Bird, D.M., Opperman, C.H., Jones S.J.M., and Baillie, D.L. 1999. The
Caenorhabditis elegans gemome: a guide in the post genomics age. Annu. Rev. Phytopathol. 37:247-265.
6. Blaxter, M. 1998.
Caenorhabditis elegans is a nematode. Science 282:2041-2046.
7. Blaxter, M. and Bird, D. 1997. Parasitic nematodes. Pp. 851-878. In: Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (eds).
C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY 1222 pp.
8. Blaxter, M.L., De Ley, P., Garey, J.R., Liu, L.X., Scheldeman, P., Vierstraete, A., Vanfleteren, J.R., Mackey, L.Y., Dorris, M., Frisse, L.M., Vida, J.T. and Thomas, W.K. 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392:71-75.
9. Burglin, T.R., Lobos, E., and Blaxter, M.L. 1998. Caenorhabditis elegans as a model for parasitic nematodes. Int. J. Parasitol. 28:395-411.
10. Chervitz, S. A. et al. 1998. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science 282:2022-2028.
11. Cowden, C., Sithigorngul, P., Brackley, P., Guastella, J., Stretton, A.O.W. 1993. Localization and differential expression of FMRFamide-like immunoreactivity in the nematode.
Ascaris suum. J. Compl. Neurol. 333:455-468.
12. Davis, E.L., Hussey, R.S., Baum, T.J., Bakker, J., Schots, A., Rosso, M-N. and Abad, P. 2000. Nematode parasitism genes. Annu. Rev. Phytopathol. 38:365-396.
13. De Giorgi, C., De Luca, F., Di Vito, M., and Lamberti, F. 1997. Modulation of expression at the level of splicing of
cut-1 RNA in the infective second-stage juvenile of the plant parasitic nematode
Meloidogyne artiellia. Mol. Gen. Genetics 253:589-598.
14. Ding, X., Shields, J., Allen, R., and Hussey, R.S. 2000. Molecular cloning and characterization of a venom allergin AG5-like cDNA from
Meloidogyne incognita. Int. J. Parasitol. 30:77-81.
15. Dong, K. and Opperman, C.H. 1997. Genetic analysis of parasitism in the soybean cyst nematode Heterodera glycines. Genetics 146:1311-1318.
16. Epstein, H. F. and Shakes, D. C. (eds) 1995.
Caenorhabditis elegans: Modern Biological Analysis of an Organism. Methods in Cell Biology. Vol. 48. Academic Press, NY 659 pp.
17. Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. and Mello, C. C. 1998. Potent and specific genetic interference by double-stranded RNA in
Caenorhabditis elegans. Nature 391:806-811.
18. Gilleard, J. S., Barry, J. D., Johnstone, I. L. 1997. Cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17:2301-2311.
19. Grant, W. N. 1992. Transformation of
Caenorhabditis elegans with genes from parasitic nematodes. Parasitol. Today 8:344-346.
20. Guo S., and K.J. Kemphues. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81:611-620.
21. Hussey, R. S. and Williamson, V. M. 1998. Physiological and molecular aspects of Nematode Parasitism. In: "Plant and Nematode Interactions". Pp. 87-108. Barker, K. R., Pederson, G. A. and Windham, G. L. (eds). Agronomy Monograph 36; ASA, CSSA, SSSA, Madison, WI. 771 pp.
22. Hussey, R. S., Davis, E. L. and Ray, C. 1994.
Meloidogyne stylet secretions. Pp. 233-249. In: Lamberti, F., De Giorgi, C., and Bird, D.M.. "Advances in molecular plant nematology". Plenum Press, NY. 309 pp.
23. Jansen, G., Hazendonk, E., Thijssen, K. L. and Plasterk, R. H. A. 1997. Reverse genetics by chemical mutagenesis in
Caenorhabditis elegans. Nature Genetics. 17:119-121.
24. Jasmer, D. P., Perryman, L. E., Conder, G. A., Crow, S. and McGuire, T. 1993. Protective immunity to
Haemonchus contortus induced by immunoaffinity isolated antigens that share a phylogenetically conserved carbohydrate gut surface epitope. J. Immunol. 151:5450-5460.
25. Koltai, H., Chejanovsky, N., Raccah, B. and Spiegel, Y. 1997. The first isolated collagen gene of the root-knot nematode
Meloidogyne javanica is developmentally regulated. Gene 196:191-199.
26. Kuwabara, P.E. and Shah, S. 1994. Cloning by synteny: Identifying C. briggsae homologues of
C. elegans genes. Nucleic Acids Res. 4414-4418.
27. Lambshead, P.J.D. 1993. Recent developments in marine benthic biodiversity research. Oceanis 19:5-24.
28. Mellow, C. and Fire, A. 1995. DNA transformation. Pp. 452-482. In: Epstein, H. F. and Shakes, D. C. (eds) 1995.
Caenorhabditis elegans: Modern Biological Analysis of an Organism. Methods in Cell Biology. Vol. 48. Academic Press, NY 659 pp.
29. Nelson, L. S. and Rosoff, M.L., and Li, C. 1998. Disruption of a neuropeptide gene,
flp-1, causes multiple behavioral defects in
Caenorhabditis elegans. Science 281:1686.
30. Niles, R.K. and Freckman, D.W. 1998. From the ground up: nematode ecology in bioassessment and ecosystem health. Pp. 65-85. In: Barker, K.R., Pederson, Gary A., and Windham, G.L. "Plant and Nematode Interactions. American Society of Agronomy, Madison, WI .
31. Pennisi, E. 1998. Worming secrets from the
C. elegans genome. Science 282:1972-1974.
32. Qin, L., Smant, G., Stokkermans, J., Bakker, J., Schots, A. and Helder, J. 1998. Cloning of a trans-spliced glyceraldehyde-3-phosphate-dehydrogenase gene from the potato cyst nematode
Globodera rostochiensis and expression of its putative promoter region in
Caenorhabditis elegans. Mol. Biochem. Parasitol. 96:59-67.
33. Ray, C., Wang, T.Y. and Hussey, R.S. 1996. Identification and characterization of the
Meloidogyne incognita col1 cuticle collagen gene. Mol. Biochem. Parasitol. 83:121-124.
34. Riddle, D. L. and Albert, P. S. 1997. Genetic and environmental regulation of dauer larva development. Pp. 739-768. In: Riddle, D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (eds).
C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY 1222 pp.
35. Riddle, D. L., Blumenthal, T., Meyer, B. J. and Priess, J. R. 1997.
C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 1222 pp.
36. Riddle, D.L. and Giorgi, L.L. 1990. Advances in research on
Caenorhabditis elegans: application to plant parasitic nematodes. Ann. Rev. Phytopathol. 28:247-269.
37. Sasser, J.N. and Freckman, D.W. 1987. A world perspective of nematology: The role of the society. Pp 7-14. In: Veech J.A. and Dickson, D.W. (ed.) "Vistas on Nematology". Soc. Nematol., Hyattsville, MD.
38. Smith, T. S., Munn, E. A., Graham, M., Tavernor, A. S., and Greenwood, C. A. 1993. Purification and evaluation of the integral membrane protein H11 as a protective antigen against
Haemonchus contortus. Int. J. Parasitol. 23:271-280.
40. The
C. elegans Genome Consortium. 1999. How the worm was won: the
C. elegans genome sequencing project. TIG 15:51-58.
41. The
C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode
C. elegans: A platform for investigating biology. Science 282:2011-2046.
42. Wood, W. B. (ed.) and the Community of
C. elegans Researchers. 1988. The Nematode
Caerorhabditis elegans. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
43. Zuckerman, B.M. and Jansson, H.B. 1984. Nematode chemotaxis and possible mechanisms of host-prey recognition. Ann. Rev. of Phytopathol. 22:95-114.
