Cassava and Its Importance in Sub-Saharan Africa
Cassava (Manihot esculenta Crantz, family Euphorbaiceae, synonyms: yucca, manioc, and mandioca), a native to South America, is believed to have been introduced into sub-Saharan Africa by the Portuguese traders during the 16th century (17). According to the Food and Agriculture Organization of the United Nations (FAO), cassava is currently the third most important source of calories in the tropics, after rice and corn, and more than 800 million people use cassava as a source of food and income generation in Africa, Asia, and Latin America. Among the cassava-growing regions of the world, Africa accounts for more than 50% of the global cassava production of 233.8 million metric tons (22). The resilience of cassava enables it to grow successfully under a wide range of agro-ecological zones where cereals and other crops cannot thrive, making it a suitable crop for poor farmers to cultivate under marginal environments in Africa. The other attraction for farmers to grow cassava is that it produces higher yields per unit of land than other crops such as yams, wheat, rice, and maize. The pivotal role of cassava in the lives of Africans is evident from many documents. For example, Flora Nwapa, a famous Nigerian novelist and poet, described how cassava sustained life in Nigeria, including during the civil war period of the late 1960s. In the book titled "Cassava Song and Rice Song" (49), she equated cassava to a mother and eloquently praised "Mother Cassava" as a staple food for the majority of "have-nots" as opposed to crops like rice, which is viewed as an expensive, imported food not affordable by the poor.
Cassava is cultivated as a tuberous root crop and its roots (Fig. 1) are the major source of dietary starch. The tubers are eaten fresh and in various forms of processed food. Cassava is grown in sub-Saharan Africa by resource-poor farmers, many of them women, as an intercrop with vegetables, plantation crops (coconut, oil palm and coffee), yams, melon, sweet potato, maize, sorghum, millet, cowpea, groundnut (= peanut), and other legumes for food security and assured household income. Cassava leaves are also consumed as a green vegetable, especially in East Africa, to provide an important source of proteins, minerals, and vitamins. Several African countries are gradually replacing wheat flour with cassava flour in the production of staples like bread and noodles. However, there is an increased awareness that excessive reliance on cassava could lead to malnutrition in these countries, since the tubers are a poor source of protein, vitamins A and E, iron, and zinc (44). In recent years, cassava has been increasingly used as raw material in the manufacture of various industrial products such as starch and flour. With increased prospects of starch from cassava as a source of ethanol for biofuels, its cultivation is transforming from subsistence to a more commercially-oriented farming enterprise (44). Consequently, cassava acreage has been increasing throughout Africa.
| |

Fig. 1. Cassava produces underground tubers (A) that are used as a source of food and for other purposes. Cassava mosaic disease (CMD) affects overall growth of the plant and production of tubers depending on the level of infection. Plants severely affected with CMD (D) show poor growth with no tubers and a moderately affected plant (C) produces few tubers with intermediate growth when compared to a healthy plant (B). |
|
Cassava Mosaic Disease
Cassava is vulnerable to a broad range of diseases caused by viruses (Table 1). Among them, cassava mosaic disease (CMD) is the most severe and widespread, limiting production of the crop in sub-Saharan Africa. CMD produces a variety of foliar symptoms that include mosaic, mottling, misshapen and twisted leaflets, and an overall reduction in size of leaves and plants (Figs. 2 and 3). CMD-affected cassava plants produce few or no tubers (Fig. 1C and 1D) depending on the severity of the disease and the age of the plant at the time of infection.
Table 1. Viruses infecting cassava (Adapted, with modifications from Thottappilly et al., 2003).
| Virus |
Genus/Family |
Symptoms |
Vector |
Distribution* |
| Cassava American latent virus (CsALV) |
Nepovirus/
Comoviridae. |
Symptomless |
Unknown |
Brazil and Guyana |
| Cassava brown streak virus (CBSV) |
Ipomovirus/
Potyviridae |
Brown, elongate necrotic lesions on stems; secondary and tertiary vein chlorosis; corky brown necrosis in tuberous roots |
Whitefly |
East and Southern Africa |
| Cassava brown streak Uganda virus |
Ipomovirus/
Potyviridae |
Brown, elongate necrotic lesions on stems; secondary and tertiary vein chlorosis; corky brown necrosis in tuberous roots |
Whitefly |
East and Southern Africa |
| Cassava Colombian symptomless virus (syn. Cassava Caribbean mosaic virus) |
tentative Potexvirus/
Flexiviridae |
Symptomless |
Unknown |
Columbia |
| Cassava common mosaic virus (CsCMV) |
Potexvirus/
Flexiviridae |
Mild mosaic |
Unknown |
South America |
| Cassava frogskin-associated virus (CFSV) |
tentative Oryzavirus/
Reoviridae |
Frog skin symptoms in tubers |
Unknown |
South America |
| Cassava green mottle virus (CsGMV) |
Nepovirus/
Comoviridae. |
Local and systemic mottle |
Unknown |
Australasia and Pacific Islands, Solomon Islands |
| Cassava Ivorian bacilliform virus (CsIBV) |
Unassigned
Ourmiavirus |
Symptomless |
Unknown |
Cote d'Ivoire |
| Cassava symptomless virus (CsSLV) |
unassigned
Nucleorhabdovirus/
Rhabdoviridae |
Symptomless |
Unknown |
Brazil |
| Cassava vein mosaic virus (CSVMV) |
Cavemovirus/
Caulimoviridae. |
Vein mosaic |
Unknown |
Brazil |
| Cassava virus C (CsVC) syn. Cassava Q virus |
Ourmiavirus/
Unassigned |
Pronounced leaf fleck |
Unknown |
Cote d'Ivoire |
| Cassava virus X (CsVX) |
Potexvirus/
Flexiviridae |
Symptomless |
Unknown |
Columbia |
| Kumi viruses A and B |
Uncharacterized |
Pronounced leaf mottle |
Unknown |
Kumi district of Uganda |
| African cassava mosaic virus (ACMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
Africa and India |
| East African cassava mosaic Cameroon virus (EACMCV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
West Africa, Tanzania |
| East African cassava mosaic Kenya virus (EACMKV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
East Africa |
| East African cassava mosaic Malawi virus (EACMMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
Malawi |
| East African cassava mosaic virus (EACMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
East Africa |
| East African cassava mosaic virus-Uganda (EACMV-UG) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
Sub-Saharan Africa |
| East African cassava mosaic Zanzibar virus (EACMZV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
Zanzibar, Madagascar |
| Indian cassava mosaic virus (ICMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
India, Sri Lanka, Togo |
| South African cassava mosaic virus (SACMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
South Africa, Malawi, Madagascar, Zimbabwe |
| Sri Lankan cassava mosaic virus (SLCMV) |
Begomovirus/
Geminiviridae |
Mosaic, leaf distortion and stunting |
Whitefly |
India and Sri Lanka |
* Based on available records.
| |

Fig. 2. Symptoms of cassava mosaic disease (CMD) on cassava. An infected plant showing severe stunting and distortion of leaves (A) compared to a healthy plant (B). Leaves of CMD-affected plants produce misshapen and twisted leaflets with mosaic and mottling symptoms (C and D). |
|
| |

Fig. 3. Cassava field with a mixture of healthy and infected plants. Cuttings from plants affected by cassava mosaic disease (arrow head) show disease symptoms soon after planting and serve as a source of inoculum for whiteflies to spread the disease to neighboring plants. |
|
Historically, the first report of CMD came from the Usambaras Mountains range in northeast Tanzania in 1894. The disease was named as "Kräuselkrankheit," a German word that translates to "rippling/crinkling illness" (80) which describes symptoms observed on affected plants. Although a virus was originally believed to be the causal agent of CMD and that its transmission occurred via whiteflies (18), it viral status was not proven until the 1970s when small, quasi-isometric, geminate particles, were found in symptomatic host tissues (32). However, it would not be until many years later that the virus, African cassava mosaic virus (ACMV), was molecularly characterized (66) and Koch’s postulates fulfilled (11).
Further molecular characterization of ACMV isolates from CMD-affected plants in Kenya later revealed the presence of a second virus with a genome organization similar to ACMV, but with distinct serological properties. This ‘new’ virus was named East African cassava mosaic virus (EACMV) (35) to reflect its geographical association, which was thought to be restricted to East Africa. Since then, nine distinct cassava mosaic viruses have been characterized worldwide from CMD-affected cassava plants and seven of them are from sub-Saharan Africa. These viruses are ACMV, EACMV, East African cassava mosaic Cameroon virus (EACMCV) (27), East African cassava mosaic Kenya virus (EACMKV) (15), East African cassava mosaic Malawi virus (EACMMV) (82), East African cassava mosaic Zanzibar virus (EACMZV) (40), and South African cassava mosaic (SACMV) (7). Two other viruses, Indian cassava mosaic virus (ICMV) (41,61) and Sri Lankan cassava mosaic virus (SLCMV) (61), were reported from the Indian sub-continent. Currently, the International Committee on Taxonomy of Viruses (ICTV) has placed all of these viruses in the genus Begomovirus, the largest genus in the family Geminiviridae, and collectively, they are also called the cassava mosaic begomoviruses (CMBs) or cassava mosaic geminiviruses (CMGs). Detailed historical accounts of CMD etiology have been reviewed previously (6,70).
Distribution of CMBs
Previously, CMBs were thought to show geographic structuring with ACMV limited to West and Central African countries towards the west of the Rift Valley and in South Africa, EACMV to the eastern part of the Rift Valley in coastal Kenya, Tanzania, Malawi, Zimbabwe and Madagascar, and ICMV to India and Sri Lanka (33). However, subsequent reports have shown that most of the seven CMBs reported from sub-Saharan Africa are widespread across the sub-continent (6,8,15,56), whereas ICMV and SLCMV appear to have remained confined to cassava-growing regions of India and Sri Lanka (56,70). Interestingly, there is no report to date of CMD from South America, the center of origin of cassava. This suggests that CMBs could be ‘native’ to Africa where they perpetuated in indigenous hosts, and that cassava became an ‘accidental’ host after its introduction into the continent. This type of ‘new encounter phenomenon’ (14), where a pathogen that co-evolved with indigenous plant species makes a host ‘jump’ to an introduced plant species where it now becomes an important pathogen, has been reported for several other crops that have been introduced to Africa (72).
Impact of CMD on Cassava
CMBs induce several morphological and cytological modifications in cassava and the experimental host Nicotiana benthamiana Domin, and many of these have been described earlier (6). The symptoms and accompanying cellular modifications depend on whether cassava is infected with a single virus, or if there is a concurrent infection of two or more CMBs resulting in synergistic interactions (27). Characteristic symptoms of CMD for plants infected with a single virus range from green to yellow mosaic of affected leaves coupled with leaf distortion (Fig. 2), regardless of the infecting CMB species. These morphological alterations in cassava often result in lost tuber yield (Fig. 1C and 1D), and significant storage root yield losses can occur even in resistant genotypes that show only mild or no foliar symptoms (62). Overall, storage root yield loss across sub-Saharan Africa were estimated between 15 and 24 % annually, which is equivalent to 12-23 million tons or an annual loss of US$ 1.2 to 2.3 billion (74).
Morphology and Genome Organization of CMBs
CMBs are characterized by their geminate particles (30 x 20 nm) and circular, bipartite, single-stranded DNA (ssDNA) genomes encapsidated in protein coats of about 30 kDa (65). The two genomic components of CMBs, referred to as DNA A and DNA B (66), are distinct in terms of the number and function of genes each encodes but both share a stretch of ~ 200 nucleotide-long sequences referred to as the common region (Fig. 4). The common region encompasses a conserved stem-loop structure and contains several regulatory elements including the nonanucleotide TAATATTA↓C sequence (arrow denotes the nicking site for initiation of virion-sense DNA replication), and the TATA box and iterons which act as binding sites for the replication-associated protein (31). By far, the most informative of both genomic components is the DNA A that encodes two overlapping virion-sense open reading frames (ORFs) AV2 and AV1, and at least four overlapping complementary-sense ORFs AC1, AC2, AC3 and AC4 (Fig. 4; 25,36,65). AV1 encodes the coat protein gene (CP) and is the determinant of vector transmission (34) in addition to its role in genome encapsidation. As depicted by their names, the complementary-sense genes AC1 through AC4, individually and in concert, are implicated in the replication of CMBs within the host cell. ORF AC1 encodes a replication-associated protein (Rep), AC2 a transcriptional activator protein (TrAP), and AC3 a replication enhancer protein (REn). ORF AC4 plays a role as a host activation protein, which serves as an important symptom determinant implicated in cell-cycle control, and may also counteract the host response to Rep gene expression (65). In a recent study, the AC4 protein of EACMCV was shown to be a pathogenicity determinant and suppressor of the systemic phase of RNA silencing in N. benthamiana (28). The AC2 of ACMV can act as a trans-activator of several plant genes (76), in addition to its role as a trans-activator of the late viral genes AV1 and BV1 (68). AC2 protein also functions in the suppression of post-transcriptional gene silencing (79). A putative ORF, AC5, which is encoded in the complimentary sense and embedded within the CP gene, has also been reported for some CMBs (35) but has not yet been proven to be transcribed and translated. The AV2 ORF, a signature of Old World begomoviruses (60), functions as a movement protein. The two ORFs of the DNA B component, BV1 and BC1, encode the nuclear shuttle protein and the movement protein, respectively (Fig. 4). These two ORFs are non-overlapping and code for genes that play a role in intra- (BV1) and inter- (BC1) cellular movement of virions within the host plant cell (36).
| |
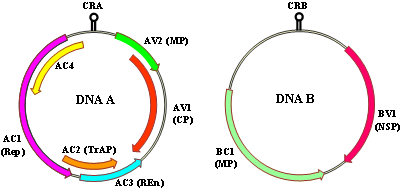
Fig. 4. Genome organization of DNA A and DNA B components of cassava-infecting begomoviruses. The genomic maps were drawn based on DNA A (GenBank Accession No. X17095) and DNA B (GenBank Accession No. X17096) sequences of African cassava mosaic virus. Open reading frames (ORFs) are denoted as either being encoded in the virion-sense (V) or complementary-sense (C) strand, preceded by component designation (A or B). CRA, common region A; CRB, common region B; CP, coat protein; MP, movement protein; Rep, replication-associated protein; TrAP, transcriptional activator protein; REn, replication enhancer protein. |
|
Recombinant Strains and Subviral Agents Associated with CMD
Perhaps no event has drawn more attention to CMD, as did the epidemic that broke out in Uganda during the early to mid 1990’s (29,53). This epidemic, characterized by a bizarre CMD symptom-type (Fig. 5), soon spread to other countries in East Africa including Kenya and Tanzania (53). Molecular characterization of the CMB associated with this unusual form of CMD revealed that it was a novel CMB that shared genomic properties with both ACMV and EACMV. This novel CMB was designated an Ugandan variant of EACMV called EACMV-UG (20), because its DNA A genome was 16% and 84% similar to that of ACMV and EACMV, respectively, as a consequence of recombination between the two parental viruses (81).
| |

Fig. 5. Cassava plant affected by an ‘unusual’ form of cassava mosaic disease (CMD) showing characteristic ‘candle stick’ symptoms as a result of infection by a recombinant cassava mosaic begomovirus known as East African cassava mosaic virus-Uganda (EACMV-UG). The CMD pandemic that occurred in East Africa during the 1990s was caused by EACMV-UG and devastated many cassava farms forcing thousands of subsistence farmers to abandon the crop (53) and leading to famine-related deaths (54). |
|
The CMD pandemic caused by EACMV-UG devastated many cassava farms and forced thousands of subsistence farmers to abandon the crop (53), and also resulted in famine-related deaths (54). EACMV-UG has since been reported from several countries in sub-Saharan Africa including Sudan (33), Rwanda (38), the Democratic Republic of Congo (48), Burundi (9) and Gabon (37). More recently, EACMV-UG has been reported from Burkina Faso (75), and Cameroon (2), thus indicating the westward movement of the virus within the African continent.
The rapid spread of this recombinant virus could be due to the indiscriminate dissemination of virus-infected vegetative cassava cuttings across sub-Saharan Africa, probably as a consequence of poor sanitation and inefficient quarantine programs in many countries of the sub-region. Although other recombinant CMBs, such as SACMV, EACMCV, EACMMV, EACMZV, EACMKV, SLCMV and ICMV have been identified, they appear to be localized in their distribution relative to EACMV-UG, indicating that EACMV-UG is perhaps better adapted than the other CMB strains found in sub-Saharan Africa. Furthermore, none of the recent disease surveys conducted in sub-Saharan African countries (5,15,64) have reported the incidence of the parental EACMV virus suggesting that this ‘wild-type’ EACMV has been overtaken in nature by its more fit recombinant strains. It is interesting to note that many of the recombinant CMBs have an EACMV lineage, whereas no recombinant from an ACMV lineage has been reported, despite its wide distribution across Africa and its frequent existence in mixed infections with other CMBs (3,51,58). Some of the factors that could contribute to molecular diversity among CMBs of an EACMV lineage were recently reviewed (56).
Adding to the complexity of the CMD situation is a recent report that disease resistance-breaking satellite DNA molecules have been found associated with CMD in Tanzania (46). Many ssDNA satellites of ~1.3 kb have been found associated with several begomovirus disease complexes and they are generally of two types, the nanovirus-like DNA 1 or alphasatellites, and the DNA B-like DNA β or betasatellites (12,45). The alphasatellites are capable of independent replication although they depend on the helper virus-encoded proteins for their movement and encapsidation, whereas the betasatellites depend on their helper virus for replication, movement and encapsidation (12,45). It was recently demonstrated that several CMBs showed contrasting and differential interactions with alpha- and beta-satellites derived from other Begomovirus species resulting in the modulation of symptom phenotypes by these satellites in N. benthamiana (57). Because the main thrust of CMD management in sub-Saharan Africa has been the development and deployment of disease resistant cultivars, the possibility that disease resistance-breaking satellite DNA molecules exist within the sub-region could further complicate the CMD situation and pose a serious threat to cassava production.
Transmission of CMD and Its Associated Viruses
Since cassava is a vegetatively propagated crop, CMBs and their DNA satellites can be transmitted through infected stem cuttings (Fig. 6A) and by grafting infected budwood onto healthy cassava plants (6 and references therein). Experimental transmission can also occur via biolistic inoculation using a gene gun (13). These procedures have been widely used to assess genotype performance against CMBs during resistance breeding programs.
| |

Fig. 6. Cassava mosaic disease (CMD) is primarily spread through the dissemination of stem cuttings (A) obtained from cassava plants affected by the disease. Secondary spread can occur within and between fields through the activities of the whitefly vector Bemisia tabaci (B). Cassava mosaic begomoviruses have also been documented from some non-cassava plant species such as Leucana leucocephala (C) and the wild relative of cultivated cassava, Manihot glaziovii (D) found growing in proximity to cassava fields. |
|
The whitefly vector, Bemisia tabaci (Hemiptera: Aleyrodidae; Fig. 6B), is mainly responsible for the secondary spread of CMBs (18,21) although other species of whitefly, such as B. afer, can also transmit CMD (55). Starvation of non-viruliferous whiteflies prior to acquisition feeding on infected cassava accelerated the acquisition of ACMV (21). Once acquired, a latent period of about 6-8 h must lapse before the whitefly is able to transmit the virus, which can thereafter be retained by an infectious whitefly for about 9 days (21). Viruliferous whiteflies require 10-30 min inoculation access periods for virus inoculation into healthy cassava plants. Under experimental conditions, ten viruliferous whiteflies are needed to achieve the optimal rate of transmission when released on a cassava plant (21), although a single whitefly is capable of virus transmission (18,21). ACMV is transtadially (18,21) but not transovarially (21) transmitted. In addition, CMBs are neither transmissible from cassava to cassava by mechanical inoculations nor through seed (67).
Detection of CMBs
The successful purification of CMBs (10) paved the way for development of antibody-based diagnoses of viruses associated with CMD. Polyclonal antibodies have been used for the detection of ACMV in cassava leaf samples by the double antibody sandwich (DAS) method of enzyme-linked immunosorbent assay (ELISA; 63) and immunosorbent electron microscopy (59). The availability of a panel of monoclonal antibodies (69) gave impetus for rapid detection and discrimination of CMBs using triple antibody sandwich-ELISA (34,69). Although diagnosis of CMBs by ELISA is versatile and can be used for large-scale testing of field samples in diagnostic surveys (52), its major limitation lies in its inability to distinguish different CMBs in mixed virus infections (70). In addition, similarities in the coat protein epitopes of recombinant CMBs such as EACMV-UG and their parentals makes it difficult for these viruses to be successfully differentiated by ELISA (70).
The advent of the polymerase chain reaction (PCR) technique has advanced molecular diagnosis of CMBs in several African countries including Cameroon, Nigeria, Tanzania, Kenya, South Africa, and so on (5,8,27,42,43,47,51,58,64). These assays were developed using oligonucleotide primers specific to the DNA A component of CMBs in most cases. Amplified DNA fragments were then analyzed using restriction enzymes (39,64), used in heteroduplex mobility assays (8) or sequenced for profiling CMBs (5,42,43,47,58,64). Recently, a multiplex PCR assay was developed for simultaneous detection of ACMV and EACMCV in CMD-affected cassava plants (3). The development of this assay has enhanced the capacity for rapid and reliable assessment of CMBs in epidemiological studies and crop improvement and phytosanitary programs in many sub-Saharan African countries. Nevertheless, ELISA is still a valuable and affordable diagnostic tool in countries where the capacity for molecular diagnostics is often lacking.
Epidemiology of CMD
Studies have shown that spread of CMD into new cassava fields from external sources can occur much more rapidly than within field spread (23), due to the nature of the whitefly vector. Also, the direction of prevailing wind and orientation of the farm could influence the extent of primary infection since the flight of viruliferous whiteflies into new plantings could be aided by wind (71). Since the population of the whitefly vector fluctuates during the growing season in different agroecologies, farmers in sub-Saharan Africa were advised to target periods of low vector populations when planting cassava in order to reduce CMD incidence during early stages of the crop (71).
Studies have also shown that immigrant whitefly populations tended to preferentially alight on plants in the outer rows of a cassava field (19,26), leading to greater CMD incidence at the field margins (23). Thus, some farmers plant CMD-resistant cassava cultivars at field margins to ‘buffer’ or slow CMD spread to more susceptible genotypes (30,53,73). In addition, climatic conditions under which cassava is grown and the ‘cultivar x environment’ interactions, have also been shown to significantly influence whitefly vector populations and subsequent spread of CMD (1).
Experimental and Alternative Hosts of CMBs
The experimental host range of CMBs was originally believed to be restricted to members of the family Solanaceae, especially those belonging to the genera Nicotiana and Datura (11). However, SLCMV has been shown to infect Ageratum conyzoides L. (family Asteraceae) and Arabidopsis thaliana (L.) Heynh. (family Brassicaceae) (70 and references therein). ACMV and EACMV have been detected in M. glaziovii Müll. Arg. (a wild species of cassava), Combretum confertum (Benth.) M.A. Lawson (Combretaceae), Senna occidentalis (L.) Link (Fabaceae), and Leucana leucocephala (Lam.) De Witt (Fabaceae), while ACMV alone has been detected in Jatropha multifida L. (Euphorbiaceae), Laportea (Fleurya) aestuans (L.) Chew (Urticaceae), soybean (Glycine max L. Merr.; Fabaceae), Ricinus communis L. (Euphorbiaceae), and suspected to infect Hewittia sublobata (L.f) Kuntze (Convolvulaceae) in Kenya and Laportea (Fleurya) aestuans in Nigeria (4,42,51,70 and references therein).
Recently, definitive evidence was provided for the occurrence of CMBs in non-cassava plant species including S. occidentalis, L. leucocephala, G. max, R. communis, C. confertum and M. glaziovii (Figs. 6C and 6D) based on comparative phylogenetic analyses of complete DNA A genomes of ACMV (Fig. 7A) and EACMCV (Fig. 7B) isolated from these plants and cassava (5). These results were corroborated by another study that provided evidence for the occurrence of ACMV and EACMV-UG in two weed species, Centrosema pubescens Benth. (Fabaceae) and Pueraria javanica (Benth.) Benth. (Fabaceae), sampled from the Democratic Republic of Congo (Fig. 7; 43). Anecdotal reports indicate that many of the non-cassava CMB-infected plant species also support high whitefly populations. Therefore, these non-cassava plants could be playing a significant role in the epidemiology of CMD by serving as an inoculum and vector source.
| |
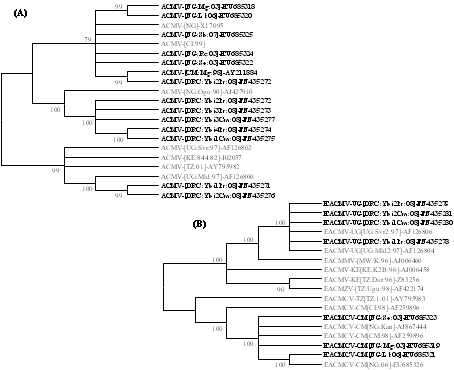
Fig. 7. Unrooted maximum likelihood (ML) phylogenetic tree depicting the evolutionary history of natural populations of (A) ACMV and (B) EACMV-type viruses obtained from cassava (regular fonts) and non-cassava plant species (in bold fonts). The ML trees were inferred based on the model specific to each of the sequence datasets and trees with the highest log likelihoods (-8084.5525 and -11536.7569 for ACMV and EACMV datasets, respectively) are shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The analyses, conducted using MEGA5 software, involved 21 and 18 complete DNA A nucleotide sequences of ACMV and EACMV-type viruses, respectively. |
|
Management of CMD
Numerous approaches have been developed for the management of CMD and many of these approaches have been mentioned in previous sections and discussed in several review articles (6,71,78). Chemical control of the whitefly vector has seldom been practiced by farmers in Africa for economic reasons. In addition, pesticides are least effective in controlling arthropod-borne viruses if the main spread is from external sources and not within crops (71). The negative impact of pesticides on the environment and risks to beneficial organisms, including natural enemies and farmers’ health, also makes pesticide use less appealing. The potential for biological control of the whitefly vector remains to be explored, although studies in this direction have been initiated recently in East Africa.
A considerable amount of effort and resources have been devoted to breeding for resistance to CMBs and their whitefly vector, especially by scientists at the International Institute of Tropical Agriculture (IITA, Ibadan, Nigeria), the International Centre for Tropical Agriculture (CIAT, Cali, Columbia), and their collaborators in national, regional and international laboratories. Such disease-resistant materials have been widely distributed across sub-Saharan Africa for use by local farmers. However, one limitation of resistant varieties lies in the fact that some of them may accommodate moderate to high levels of virus (50). Such varieties could actually be considered tolerant, rather than resistant, to virus infection because they permit virus replication although visible symptoms are not apparent. From an epidemiological standpoint, such materials could serve as sources of virus inoculum if conditions for vector spread are favorable. In addition, the possibility that disease resistance-breaking satellite DNA molecules exist in nature in sub-Saharan Africa (as discussed above) jeopardizes true resistant cassava cultivars that have been widely distributed across the region.
Recently, global efforts have focused on the development of transgenic cassava cultivars resistant to CMD (56,77,78). Such efforts are now being integrated into the larger goal of the BioCassava Plus initiative, funded by the Bill and Melinda Gates Foundation, which has the main objective of reducing malnutrition by delivering improved cassava cultivars that provide complete and balanced nutrition in a readily marketable and higher yielding food crop (http://biocassavaplus.org). It is hoped that the recent completion of the draft genome of cassava by a consortium of international scientists would give a boost to these efforts. Although transgenic resistance against CMD has shown some promising results under controlled conditions (78), their performance under field conditions where there is high disease pressure and a greater abundance of vectors remains to be evaluated.
Among the various strategies evaluated for CMD management, planting virus-free cuttings is the most effective for minimizing spread of the disease in Africa (24, 71). However, when it comes to effective management of CMD, it is impracticable to point at a ‘one-size-fits-all’ strategy. As such, management efforts should integrate different approaches (‘traditional’ vs. ‘novel’; ‘ancient’ vs. ‘modern’; ‘conventional’ vs. ‘unconventional’), which will hold better promise for comprehensive CMD management in the region. Also, the potential roles of volunteer cassava plants and alternative hosts of CMBs (discussed above) in the evolution of CMBs and subsequent development of CMD have been largely overlooked, which may impact the success of any management strategy.
Conclusions and Perspectives
Despite the many advances made since CMD was first reported over a century ago, CMD continues to be a significant threat to sustainable cassava production in sub-Saharan Africa. Although molecular studies on CMBs and host-virus-vector interactions are advancing our knowledge on various aspects of CMD, there are still many questions that need to be addressed for mitigating the negative impact of the disease on cassava production. One of the key gaps in knowledge on the epidemiology of CMD in sub-Saharan Africa is to define the role played by non-cassava plant species and alternative/reservoir hosts in the perpetuation of CMBs. Since cassava was introduced from South America in the 16th Century, it is likely that CMBs endemic to Africa that infected indigenous African plant species became adapted to cassava. Thus, it is plausible that native plant species could act as alternative and/or reservoir hosts for CMBs and contribute to continued virus evolution and future disease epidemics. Since cassava is a vegetatively propagated crop, distribution of infected vegetative cuttings largely contributes to long distance dissemination of CMBs. Hence, concerted efforts are needed to enforce quarantine regulations in African countries to prevent dissemination of CMBs by propagation materials. A prerequisite to prevention is the availability of sensitive, reliable and rapid diagnostic tools for detection and differentiation of CMBs, including the recently developed multiplex PCR assay for detecting multiple CMBs in mixed-infected plants (3). Such assays will need to be constantly updated to detect newly evolving CMBs, as well as to incorporate detection of other important cassava viruses such as Cassava brown streak virus (genus Ipomovirus; family Potyviridae) in ‘clean plant’ programs. The intensification of research efforts and the achievement of a synergy between traditional and novel approaches for CMD management will provide a holistic approach towards making cassava improvement an integral part of poverty alleviation in sub-Saharan Africa. In this regard, recent efforts to build and improve capabilities of scientists in the region to address these challenges within local boundaries, such as the current initiatives in eastern and central Africa (16), are a welcome development.
Acknowledgments
The review article, based on a chapter in the Ph.D. dissertation of the first author, is dedicated to the memory of the late Dr. Francis O. Ogbe for his contributions to research on cassava mosaic disease. The authors are grateful for funding from the USAID-Linkage Grant through the International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria. PPNS# 0570, Department of Plant Pathology, College of Agricultural, Human, and Natural Resource Sciences Agricultural Research Center Project No. WNPO 0616, Washington State University, Pullman, WA 99164-6240, USA. We thank Dr. Richard Larsen (USDA-ARS IAREC, Prosser, WA) and Ranajit Bandyopadhyay (IITA, Ibadan, Nigeria) for reviewing the manuscript. We also thank the Early Career Professionals Committee of the American Phytopathological Society (APS) and the APS for selecting the first author as an Awardee of the Schroth Faces of the Future in Plant Virology at the 2010 Annual Meeting of the Society in Charlotte, NC.
References
1. Abdullahi, I., Atiri, G. I., Dixon, A. G. O., Winter, S., and Thottappilly, G. 2003. Effects of cassava genotype, climate and the Bemisia tabaci vector population on the development of African cassava mosaic geminivirus (ACMV). Acta Agron. Hung. 51:37-46.
2. Akinbade, S. A., Hanna, R., Nguenkam, A., Njukwe, E., Fotso, A., Doumtsop, A., Ngeve, J., Tenku, S. T. N., and Kumar, P. L. 2010. First report of the East African cassava mosaic virus-Uganda (EACMV-UG) infecting cassava (Manihot esculenta) in Cameroon. New Disease Reports 21:22. doi:10.5197/j.2044-0588.2010.021.022.
3. Alabi, O. J., Kumar, P. L., and Naidu, R. A. 2008. Multiplex PCR method for the detection of African cassava mosaic virus and East African cassava mosaic Cameroon virus in cassava. J. Virol. Methods 154:111-120.
4. Alabi, O. J., Ogbe, F. O., Bandyopadhyay, R., Dixon, A. G. O., Hughes, J., and Naidu, R. A. 2007. The occurrence of African cassava mosaic virus and East African cassava mosaic Cameroon virus in natural hosts other than cassava in Nigeria (Abstr.). Phytopathology 97:S3.
5. Alabi, O. J., Ogbe, F. O., Bandyopadhyay, R., Kumar, P. L., Dixon, A. G. O., Hughes, J. d’A., and Naidu, R. A. 2008. Alternate hosts of African cassava mosaic virus and East African cassava mosaic Cameroon virus in Nigeria. Arch. Virol. 153:1743-1747.
6. Atiri, G. I., Ogbe, F. O., Dixon, A. G. O., Winter, S., and Ariyo, O. 2004. Status of cassava mosaic virus diseases and cassava begomoviruses in sub-Saharan Africa. J. Sustainable Agric. 24:5-35.
7. Berrie, L. C., Palmer, K. E., Rybicki, E. P., and Rey, M. E. C. 1998. Molecular characterisation of a distinct South African cassava infecting geminivirus. Arch. Virol. 143:2253-2260.
8. Berry, S., and Rey, M. E. C. 2001. Molecular evidence for diverse populations of cassava infecting begomoviruses in southern Africa. Arch. Virol. 146:1795-1802.
9. Bigirimana, S., Barumbanze, P., Obonyo, R., and Legg, J. P. 2004. First evidence for the spread of East African cassava mosaic virus-Uganda (EACMV-UG) and the pandemic of severe cassava mosaic disease to Burundi. Plant Pathol. 53:231.
10. Bock, K. R., Guthrie, E. J., Meredith, G., and Barker H. 1977. RNA and protein components of maize streak and cassava latent viruses. Annals of Applied Biology 85:305-308.
11. Bock, K. R., and Woods, R. D. 1983. Etiology of African cassava mosaic disease. Plant Dis. 67:994-995.
12. Briddon, R. W., Brown, J. K., Moriones, E., Stanley, J., Zerbini, M., Zhou, X., and Fauquet, C. M. 2008. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch. Virol. 153:763-781.
13. Briddon, R. W., Liu, S., Pinner, M. S., and Markham, P. G. 1998. Infectivity of African cassava mosaic virus clones to cassava by biolistic inoculation. Arch. Virol. 143:2487-2492.
14. Buddenhagen, I. W. 1977. Resistance and vulnerability of tropical crops in relation to their evaluation and breeding. Ann. N. Y. Acad. Sci. 287:309-326.
15. Bull, S. E., Briddon, R. W., Sserubombwe, W. S., Ngugi, K., Markham, P. G., and Stanley, J. 2006. Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87:3053-3065.
16. Bull, S. E., Ndunguru, J., Gruissem, W., Beeching, J. R., and Vanderschuren, H. 2011. Cassava: constraints to production and the transfer of biotechnology to African laboratories. Plant Cell Rep. doi:10.1007/s00299-010-0986-6.
17. Carter, S. E., Fresco, L. O., Jones, P. G., and Fairbairn, J. N. 1995. Introduction and diffusion of cassava in Africa. Intl. Inst. Trop. Agric. Res. Guide No. 49.
18. Chant, S. R. 1958. Studies on the transmission of cassava mosaic virus by Bemisia spp. (Aleyrodidae). Ann. Appl. Biol. 46:210-215.
19. Colvin, J., Fishpool, L. D. C., Fargette, D., Sherington, J., and Fauquet, C. M. 1998. Bemisia tabaci (Hemiptera: Aleyrodidae) trap catches in a cassava field in Cote d’Ivoire in relation to environmental factors and the distribution of the African cassava mosaic disease. Bull. Entomol. Res. 88:369-378.
20. Deng, D., Otim-Nape, W. G., Sangare, A., Ogwal, S., Beachy, R. N., and Fauquet, C. M. 1997. Presence of a new virus closely related to East African cassava mosaic geminivirus, associated with cassava mosaic outbreak in Uganda. Afr. J. Root Tuber Crops 2:23-28.
21. Dubern, J. 1994. Transmission of African cassava mosaic geminivirus by the whitefly (Bemisia tabaci). Trop. Sci. 34:82-91.
22. FAOSTAT 2009. FAOSTAT. Available at http://faostat.fao.org (accessed 26 May 2009; verified 24 May 2011). Food and Agriculture Organization (FAO) of the United Nations, Rome, Italy.
23. Fargette, D., Fauquet, C., Grenier, E., and Thresh, J. M. 1990. The spread of African cassava mosaic virus into and within cassava fields. J. Phytopathol. 130:289-302.
24. Fargette, D., Jeger, M., Fauquet, C., and Fishpool, L. D. C. 1994. Analysis of temporal disease progress of African cassava mosaic virus. Phytopathology 84:91-98.
25. Fauquet, C. M., Briddon, R. W., Brown, J. K., Moriones, E., Stanley, J., Zerbini, M., and Zhou, X. 2008. Geminivirus strain demarcation and nomenclature. Arch. Virol. 153:783-821.
26. Fishpool, L. D. C., Fauquet, C. M., Fargette, D., Thouvenel, J. C., Burban, C., and Colvin, J. 1995. The phenology of Bemisia tabaci (Homoptera: Aleyrodidae) populations on cassava in southern Cote d’Ivoire. Bull. Entomol. Res. 85:197-207.
27. Fondong, V. N. F., Pita, S., Rey, M. E. C., de Kochko, A., Beachy, R. N., and Fauquet, C. M. 2000. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81:287-297.
28. Fondong, V. N., Reddy, R. V., Lu, C., Hankoua, B., Felton, C., Czymmek, K., and Achenjang, F. 2007. The consensus N-Myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant-Microbe Interact. 20:380-391.
29. Gibson, R. W., Legg, J. P., and Otim-Nape, G. W. 1996. Unusually severe symptoms are a characteristic of the current epidemic of mosaic virus disease of cassava in Uganda. Ann. Appl. Biol. 128:479-490.
30. Hahn, S. K., Terry, E. R., and Leuschner, K. 1980. Breeding cassava for resistance to cassava mosaic disease. Euphytica 29:673-683.
31. Hanley-Bowdoin, L., Settlage, S. B., Orozco, B. M., Nagar, S., and Robertson, D. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. CRC Crit. Rev. Plant Sci. 18:71-106.
32. Harrison, B. D., Barker, H., Bock, K. R., Guthrie, E. J., Meredith, G., and Atkinson, M. 1977. Plant viruses with circular single-stranded DNA. Nature 270:760-762
33. Harrison, B. D., Liu, Y. L., Zhou, X., Robinson, D. J., Calvert, L., Munoz, C., and Otim-Nape, G. W. 1997. Properties, differentiation and geographical distribution of geminivirus that cause cassava mosaic disease. Afr. J. Root Tuber Crops 2:19-22.
34. Harrison, B. D., Swanson, M. M., and Fargette, D. 2002. Begomovirus coat protein: serology, variations and functions. Physiol. Mol. Plant Pathol. 60:257-271.
35. Hong, Y. G., Robinson, D. J., and Harrison, B. D. 1993. Nucleotide sequence evidence for the occurrence of three distinct whitefly-transmitted geminiviruses in cassava. J. Gen. Virol. 74:2437-2443.
36. Hull, R. 2002. Genome organization. Pages 171-224 in: Matthew's Plant Virology, 4th Edn. R. Hull, ed. Academic Press, London, UK.
37. Legg, J. P., Ndjelassili, F., and Okao-Okuja, G. 2004. First report of cassava mosaic disease and cassava mosaic geminiviruses in Gabon. Plant Pathol. 53:232.
38. Legg, J. P., Okao-Okuja, G., Mayala, R., and Muhinyuza, J. B. 2001. Spread into Rwanda of the severe cassava mosaic virus disease pandemic and associated Ugandan variant of East African cassava mosaic virus (EACMV-UG). Plant Pathol. 50:796.
39. Legg, J. P., and Thresh, J. M. 2001. Cassava virus diseases in Africa. Pages 517-552 in: Proceedings of Plant Virology in Sub-Saharan Africa. J. d’A. Hughes and B. O. Odu, eds. Intl. Inst. Trop. Agric., Ibadan, Nigeria.
40. Maruthi, M. N., Seal, S., Colvin, J., Briddon, R. W., and Bull, S. E. 2004. East African cassava mosaic Zanzibar virus - a recombinant begomovirus species with a mild phenotype. Arch. Virol. 149:2365-2377.
41. Matthew, A. V., and Muniyappa, V. 1992. Purification and characterization of Indian cassava mosaic virus. Phytopathology 135:299-308.
42. Mgbechi-Ezeri, J., Alabi, O. J., Naidu, R. A., and Kumar, P. L. 2008. First report of the occurrence of African cassava mosaic virus in a mosaic disease of soybean in Nigeria. Plant Dis. 92:1709.
43. Monde, G., Walangululu, J., Winter, S., and Bragard, C. 2010. Dual infection by cassava begomoviruses in two leguminous species (Fabaceae) in Yangambi, Northeastern Democratic Republic of Congo. Arch. Virol. 155:1865-1869.
44. Nassar, N., and Ortiz, R. 2010. Breeding cassava to feed the poor. Pages 78-84 in: Scientific American (http://www.scientificamerican.com/article.cfm?id=breeding-cassava), May 2010 issue.
45. Nawaz-ul-Rehman, M. S., and Fauquet, C. M. 2009. Evolution of geminiviruses and their satellites. FEBS Lett 583:1825-1832.
46. Ndunguru, J., Fofana, B., Legg, J. P., Chellappan, P., Taylor, N., Aveling, T., Thompson, G., and Fauquet, C. 2008. Two novel satellite DNAs associated with bipartite cassava mosaic begomoviruses enhancing symptoms and capable of breaking high virus resistance in a cassava landrace. Page 141 in: Book of Abstracts, Global Cassava Partnership-First Scientific Meeting: Cassava Meeting the challenges of the New Millennium. Ghent University, Ghent, Belgium.
47. Ndunguru, J., Legg, J. P., Aveling, T. A. S., Thompson, G., and Fauquet, C. M. 2005. Molecular biodiversity of cassava begomoviruses in Tanzania: evolution of cassava geminiviruses in Africa and evidence for East Africa being a center of diversity of cassava geminiviruses. Virol. J. 2:21.
48. Neuenschwander, P., Hughes, J. d'A., Ogbe, F., Ngatse, J. M., and Legg, J. P. 2002. The occurrence of the Uganda Variant of East African cassava mosaic virus (EACMV-Ug) in western Democratic Republic of Congo and the Congo Republic defines the westernmost extent of the CMD pandemic in East/Central Africa. Plant Pathol. 51:384.
49. Nwapa, F. 1986. Cassava song and rice song. Tana Press, Enugu, Nigeria.
50. Ogbe, F. O., Atiri, G. I., Dixon, A. G. O., and Thottappilly, G. 2003. Symptom severity of cassava mosaic disease in relation to concentration of African cassava mosaic virus in different cassava genotypes. Plant Pathol. 52:84-91.
51. Ogbe, F. O., Dixon, A. G. O., Hughes, J. d’A., Alabi, O. J., and Okechukwu, R. 2006. Status of cassava begomoviruses and their new natural hosts in Nigeria. Plant Dis. 90:548-553.
52. Ogbe, F. O., Legg, J., Raya, M. D., Muimba-kankolongo, A., Theu, M. P., Kaitisha, G., Phiri, N. A., and Chalwe, A. 1997. Diagnostic survey of cassava mosaic viruses in Tanzania, Malawi and Zambia. Roots 4:12-15.
53. Otim-Nape, G. W., Bua, A., Thresh, J. M., Baguma, Y., Ogwal, S., Semakula, G. N., Acola, G., Byabakama, B., and Martin, A. 1997. Cassava mosaic virus disease in Uganda: The current pandemic and approaches to control. NARO, NRI, ODA.
54. Otim-Nape, G. W., Thresh, J. M., and Shaw, M. W. 1998. The incidence and severity of cassava mosaic virus disease in Uganda: 1990-1922. Trop. Sci. 38:25-37.
55. Palaniswami, M. S., Nair, R. R., Pillai, K. S., and Thankappan, M. 1996. Whiteflies on cassava and its role as vector of cassava mosaic in India. J. Root Crops 22:1-8.
56. Patil, B. L., and Fauquet, C. M. 2009. Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol. Plant Pathol. 10:685-701.
57. Patil, B. L., and Fauquet, C. M. 2010. Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. J. Gen. Virol. 91:1871-1882.
58. Pita, J. S., Fondong, V. N., Sangaré, A., Kokora, R. N. N., and Fauquet, C. M. 2001. Genomic and biological diversity of the African cassava geminiviruses. Euphytica 120:115-125.
59. Roberts, I. M., Robinson, D. J., and Harrison, B. D. 1984. Serological relationships and genome homologies among geminiviruses. J. Gen. Virol. 65:1723-1730.
60. Rybicki, E. P. 1994. A phylogenetic and evolutionary justification for 3 genera of Geminiviridae. Arch. Virol. 139:49-77.
61. Saunders, K., Nazeera, S., Mali, V. R., Malathi, V. G., Briddon, R., Markham, P. G., and Stanley, J. 2002. Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293:63-74.
62. Seif, A. A. 1982. Effect of cassava mosaic virus on yield of cassava. Plant Dis. 66:661-662.
63. Sequeira, J. C., and Harrison, B. D. 1982. Serological studies on cassava latent virus. Ann. Appl. Biol. 101:33-42.
64. Sserubombwe, W. S., Briddon, R. W., Baguma, Y. K., Ssemakula, G. N., Bull, S. E., Bua, A., Alicai, T., Omongo, C., Otim-Nape, G. W., and Stanley, J. 2008. Diversity of begomoviruses associated with mosaic disease of cultivated cassava (Manihot esculenta Crantz) and its wild relative (Manihot glaziovii Mull. Arg.) in Uganda. J. Gen. Virol. 89:1759-1769.
65. Stanley, J., Bisaro, D. M., Briddon, R. W., Brown, J. K., Fauquet, C. M., Harrison, B. D., Rybicki, E. P., and Stenger, D. C. 2005. Geminiviridae. Pages 301-326 in: Virus Taxonomy, VIIIth Report of the ICTV. C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball, eds. Elsevier/Academic Press, London.
66. Stanley, J., and Gay, M. R. 1983. Nucleotide sequences of cassava latent virus DNA. Nature 301:260-262.
67. Storey, H. H., and Nichols, R. F. W. 1938. Studies of the mosaic disease of cassava. Ann. Appl. Biol. 25:790-806.
68. Sunter, G., and Bisaro, D. M. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331.
69. Thomas, J. E., Massalski, P. R., and Harrison, B. D. 1986. Production of monoclonal antibodies to African cassava mosaic virus and differences in their reactivity with other whitefly-transmitted geminiviruses. J. Gen. Virol. 67:2739-2748.
70. Thottappilly, G., Thresh, J. M., Calvert, L. A., and Winter S, 2003. Cassava. Pages 107-165 in: Virus and virus-like diseases of major crops in developing countries. G. Loebenstein and G. Thottappilly, eds. Kluwer Academic Publ., Dordrecht, The Netherlands.
71. Thresh, J. M., and Cooter, R. J. 2005. Strategies for controlling cassava mosaic virus disease in Africa. Plant Pathol. 54:587-614.
72. Thresh, J. M., and Fargette, D. 2001. The epidemiology of African plant viruses: basic principles and concepts. Pages 61-111 in: Proceedings of Plant Virology in Sub-Saharan Africa. J. d’A. Hughes and B. O. Odu, eds. Intl. Inst. Trop. Agric., Ibadan, Nigeria.
73. Thresh, J. M., Otim-Nape, G. W., and Jennings, D. L. 1994. Exploiting resistance to African cassava mosaic virus. Aspects Appl. Biol. 39:51-60.
74. Thresh, J. M., Otim-Nape, G. W., Legg, J. P., and Fargette, D. 1997. African cassava mosaic virus disease: The magnitude of the problem. Afr. J. Root Tuber Crops 2:13-18.
75. Triendrebeogo, F., Lefeuvre, P., Hoareau, M., Traoore, E. V. S., Barro, N., Reynaud, B., Traore, A. S., Konate, G., Traore, O., and Lett, J. M. 2009. Occurrence of East African cassava mosaic virus-Uganda (EACMV-UG) in Burkina Faso. Plant Pathol. 58:783.
76. Trinks, D., Rajeswaran, R., Shivaprasad, P. V., Akbergenov, R., Oakeley, E. J., Veluthambi, K., Hohn, T., and Pooggin, M. M. 2005. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79:2517-2527.
77. Vanderschuren, H., Alder, A., Zhang, P., and Gruissem, W. 2009. Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 70:265-272.
78. Vanderschuren, H., Stupak, M., Futterer, J., Gruissem, W., and Zhang, P. 2007. Engineering resistance to geminiviruses - review and perspectives. Plant Biotechnol. J. 5:207-220.
79. Vanitharani, R., Chellappan, P., Pita, J. S., and Fauquet, C. M. 2004. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78:9487-9498.
80. Warburg, O. 1894. Die Kulturpflantzen Usambaras. Mittcilungen aus den Deutschen Schutzgebieten 7:131.
81. Zhou, X., Liu, Y., Calvert, L., Munoz, C., Otim-Nape, G. W., Robinson, D. J., and Harrison, B. D. 1997. Evidence that DNA A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by inter-specific recombination. J. Gen. Virol. 78:2101-2111.
82. Zhou, X., Robinson, D. J., and Harrison, B. D. 1998. Types of variation in DNA A among isolates of East African cassava mosaic virus from Kenya, Malawi and Tanzania. J. Gen. Virol. 79:2835-2840.
Additional Resources
See an Estonian translation at the 1800flowers.com blog.
