
Cucumber mosaic
Cucumber mosaic virus
Over 1200 species in over 100 families of monocots and dicots, including many vegetables, ornamentals and woody and semi-woody plants.


Figures 1 and 2
Symptoms and Signs
Symptoms
CMV infects 1200 species in over 100 plant families and can cause significant economic losses in many vegetable and horticultural crops. CMV causes a systemic infection in most host plants, but may remain symptomless in some crops like alfalfa. Symptoms of cucumber mosaic can vary greatly depending on the crop infected and the age of the plant when infection occurs.
Cucurbits: Almost all cucurbits are susceptible to CMV, with symptoms varying in severity (Figure 3A-E). Severe epinasty, downward bending of the petiole and leaf surface along with leaf reduction, are common in early season infection of summer squash (Figure 3A).

Figure 3 A-E. CMV infection of cucurbits |
Plants infected early in the season are severely stunted and leaves are malformed, and fruit are unmarketable because of pronounced rugosity (roughness) on the fruit surface, as shown on the infected zucchini plant and fruit in Figure 3B. Infection of vining crops, such as muskmelon, show severely stunted growing tips (Figure 3C), and although fruit may not show symptoms they are of poor quality. If the yellow squash variety grown lacks the precocious gene, color breaking will occur on the fruit, causing the fruit to show green blotchy patterns, but these symptoms are absent in yellow squash varieties with the precocious gene (Figure 3D). Color breaking on fruit of varieties without the precocious gene will also occur with Watermelon mosaic potyvirus (WMV) infection; however this protection does not hold true for Papaya ringspot potyvirus or Zucchini yellow mosaic potyvirus, where both foliage and fruit of yellow squash are severely affected. Pumpkin is another cucurbit, that when infected at any early stage, will express severe foliar mosaic and the fruit will show a mosaic pattern and would be unmarketable (Figure 3E).

Figure 4 A-I. CMV symptoms on non-cucurbit crops |
Pepper: Foliar symptoms of pepper plants vary with stage of infection. The initial flush of systemic symptoms typically includes a chlorosis of young leaves that may occur over the basal portion of the leaf or over the entire leaf. Oak leaf and ringspot patterns may develop on these leaves as the plant ages (Figure 4A). As new leaves emerge, these leaves develop a chlorotic mosaic pattern that tends to encompass the entire leaf. Leaves that develop subsequent to those expressing the chlorosis and chlorotic mosaic symptoms may have varied degrees of deformation including sunken interveinal lamina with protruding primary veins. These leaves also have a dull light green appearance as opposed to the dark green, rather shiny leaves of healthy pepper plants (Figure 4B). These symptom patterns vary in severity depending on the age of the plant at the time of infection with more severe symptoms typically resulting when plants are young at the time of infection. CMV infected plants also tend to be stunted with those plants infected early in development possibly expressing severe stunting, whereas plants infected at later stages of development may have little, if any, stunting. Pepper fruit may develop ringspotting and roughness leading to unmarketable fruit (Figure 4C).
Spinach: CMV infection of spinach is often referred to as spinach blight. The symptoms can vary depending upon the variety, plant age when infected, temperature, and virus strain. Typical symptoms include leaf chlorosis (Figure 4D), which can progress to cause severe blighting of the growing point and eventual plant death. In addition to chlorotic mottle, leaves can show narrowing, wrinkling with vein distortion, and inward leaf roll.
Lettuce: Symptoms of CMV infection of lettuce consist of leaf mottling, severe roughness of the leaf and occasional necrosis within the leaf tissue (Figure 4E). Plants are usually stunted if infected at an early stage of development.
Celery: CMV infection of celery is referred to as southern mosaic virus in the older literature following its description in southern production areas. Initially leaves will develop veinclearing and mosaic, and later these same leaves may show yellowing and veinal necrosis (Figure 4F). The petioles of these plants show elongated, brown to translucent, sunken beige colored lesions (Figure 4G), making the celery stalk unmarketable. Symptoms may also be transient, such that later in the season the symptoms may be limited to a dull cast, while under cool growing conditions the obvious symptoms may be muted.
Tomato: Tomato plants infected with CMV in the early stages are yellow, bushy and considerably stunted. The leaves may show a mottle similar to that caused by Tomato mosaic virus (ToMV). The most characteristic symptom of CMV is filiformity or shoestring-like leaf blades (Figures 4H and 5).

Figure 5. “Shoestringing" of CMV infected tomato |
This could be confused with a symptom of ToMV known as "fern-leaf" in which the blade of the leaflet is not as completely suppressed as in a "shoestring" leaflet, but is abnormally long and narrow. The symptoms caused by CMV can be transitory, with the bottom leaves or newly developed top leaves showing severe symptoms, while the middle leaves may appear almost normal. Severely affected plants produce few fruit, which are usually small, often mottled or necrotic, with delayed maturity.
CMV symptoms in tomato have two distinct forms depending on growing conditions.
In the greenhouse. CMV-infected tomato plants grown under greenhouse conditions may express a general chlorosis followed by an easily observed mosaic symptom on young, systemically infected leaves. These symptoms tend to be somewhat transient as newly emerging leaves may be smaller than normal and show varying degrees of deformation such as shoestringing. Internode elongation is reduced leading to overall stunting but a bushy growth habit may not occur.
In the field. The occurrence of mosaic symptoms on young, systemically-infected leaves typically include initial chlorosis and mosaic. If plants are young at the time of infection, the mosaic symptoms are rather transient due to severe leaf deformation, and in particular, an upward cupping or rolling of leaves. If plants are somewhat more mature at the time of infection, mosaic symptoms may be more obvious, in part, because such leaves are not masked by severe leaf deformation and cupped or rolled leaves. Tomato plants that are young at the time of infection remain small, sometimes severely stunted, with a bushy appearance.
Bean: Symptoms of CMV infection in bean consist of leaf curl, green mottle and blistering, and a zipperlike roughness along the main veins involving only a few leaves (Figure 4I). Foliar symptoms are most obvious and pod infection and loss is greatest when plants are infected before bloom. Early infected plants may yield no or few pods because CMV caused flower abortion and abnormal development. The pods are mostly curved, mottled and reduced in size. Plants may recover and resume normal growth with limited yield loss if plants are infected after bloom. Because symptoms can vary among varieties and can be confused with those caused by Bean common mosaic virus, it is best not to rely upon symptoms to diagnosis the particular virus(es) involved. Rather the serological technique called ELISA (enzyme-linked immunosorbent assay) should be used to confirm if the plant is infected with CMV or another virus.
Pathogen Biology
CMV is the type species of the genus Cucumovirus in the family Bromoviridae. It consists of three spherical particles, each approximately 28 nm in diameter. The CMV genome consists of three single-stranded, messenger-sense RNA molecules, designated RNA 1 (~3,350 nucleotides), RNA 2 (~3,050 nucleotides) and RNA 3 (~2,200 nucleotides) (Figure 6). Each RNA molecule is enclosed within a protective protein coat with each being a distinct single spherical-shaped particle. Thus, a mature CMV consists of three spherical particles, one particle containing RNA 1, another containing RNA 2 and the third containing RNA 3. The RNA 3 particle may contain a fourth RNA strand, referred to as RNA 4 (~1,030 nucleotides), which encodes the coat protein gene and from which the CMV coat protein is produced. This type of translational strategy, referred to as subgenomic RNA, consists of a separate strand of RNA produced during replication. While CMV RNA 3 contains the coat protein gene, that gene is only translated to produce the coat protein from its subgenomic strand. The general replication/translation scheme for RNAs 1-3 is shown in Figure 7 A-C.
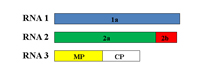
Figure 6. The three genomic RNAs of CMV |
RNA 1 encodes a single protein, referred to as 1a, shown to be necessary for replication of the viral genome (Figure 7A). The 1a protein has two functional domains: the N-terminal and C-terminal domains. The N-terminal domain has methyltransferase activity for adding a cap structure to the 5’-terminus of genomic and subgenomic RNAs, while the C-terminal domain of the 1a protein is a putative helicase which functions to “unwind” the double stranded RNA that develops during viral replication.
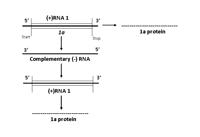
Figure 7A. Replication of RNA 1 |
RNA 2 encodes two proteins, referred to as 2a and 2b (Figure 7B). The 2a protein is N-proximal to the 2b protein and is involved in viral genome replication. The 2a protein is required for CMV replication and has characteristic RNA-dependent RNA polymerase motifs, i.e., it uses a RNA strand as a template to produce a complimentary RNA strand. The 2a protein was also shown to interact with the 1a protein (also a replication protein) in yeast two-hybrid tests. The 2b protein is translated from a separate (subgenomic) RNA strand, referred to as RNA 4A. The 2b protein counters the host’s posttranscriptional gene silencing mechanism. When CMV infects a plant, the plant may respond by activation of a resistance mechanism that essentially stops or significantly inhibits the virus from continuing its movement into, and infection of, new tissues. This particular type of resistance is referred to as gene silencing. The CMV 2b protein inhibits the ability of the plant’s gene silencing signal to initiate gene silencing in distant tissues, thereby allowing CMV to continue to invade and infect young developing tissues.
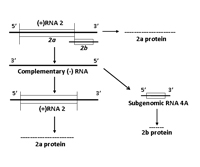
Figure 7B. Replication of RNA 2 |
RNA 3 encodes two proteins, referred to as 3a and coat protein (Figure 7C). The 3a protein (MP or movement protein in Figure 6) is essential for virus cell-to-cell movement and mutations in this protein identified host-specific differences in movement efficiency. The 3a protein was also shown to be associated with intraplant long distance movement. The coat protein is expressed from a subgenomic RNA species referred to as RNA 4. The coat protein is the only protein associated with virus particles and is the sole determinant for transmission by aphid vectors. The coat protein is involved in virus cell-to-cell movement, although suggested to have an indirect role in this movement process, and is directly involved in intraplant long distance movement. It should be noted that mutations in 1a, 2a and 2b genes affected virus movement in some hosts.
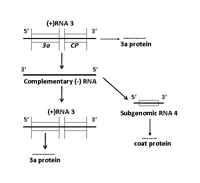
Figure 7C. Replication of RNA 3 |
In addition to the CMV genomic and subgenomic RNA species, some strains of CMV support a satellite RNA (designated RNA 5 or satRNA). The satRNA is a single-stranded molecule of approximately 332 to 342 nucleotides in length and is completely dependent on CMV for its replication. Moreover, satRNA is encapsidated in CMV particles, which allows spread from plant to plant along with CMV, by aphid vectors. The satRNA, however, does not provide CMV (i.e., the helper virus) with any essential function. The occurrence of satRNA with CMV may have no effect on CMV-induced symptoms or may intensify symptoms by production of systemic chlorosis or necrosis or may actually attenuate symptoms typically induced by CMV. The effect on disease expression is dependent on interaction of the satRNA species, the isolate or strain of CMV and the host plant species and variety. For example, a severe lethal necrosis in tomato occurred due to one CMV-satRNA interaction that affected thousands of acres of processing tomatoes in southern Italy (Figure 8).
CMV replicates in the cytoplasm following a characteristic strategy used by single-stranded messenger-sense viral genomes, whereby viral genomic RNA molecules serve as templates for generation of non-translated minus-sense RNA molecules complementary in sequence to the respective genomic RNA species. Genomic and subgenomic RNAs are generated from the respective complementary minus-sense RNA species. Proteins are translated from specific viral genes in the cytoplasm. Cell-to-cell movement occurs through plasmodesmata and intraplant long distance movement occurs through the phloem.
CMV strains are divided into two subgroups, designated subgroups I and II, that are distinguished by serological relationships and sequence analysis. Subgroup I strains are further divided into IA and IB, based on differences in pathogenicity in cowpea (Vigna unguiculata), whereby IA strains induce systemic mosaic symptoms and IB strains induce necrotic local lesions on inoculated leaves. Some CMV strains are host specific, infecting certain hosts in the same family like the legume strain of CMV. CMV is serologically related to Tomato aspermy virus and Peanut stunt virus.

Figure 8. CMV-satRNA interaction |
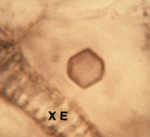
Figure 9. “Empty” virus crystal |
'Empty' virus crystals lacking virions located in a vacuole may appear as a neat hexagonal outline (Figure 9), but more often appear as 'empty' circles. Such 'empty' crystals are diagnostic for CMV, but not for cucumoviruses in general.
Disease Cycle and Epidemiology
Disease Cycle and Epidemiology
Because of its wide host range, numerous weeds can serve as reservoirs for CMV and contribute to virus spread to crops at the beginning of the season. Perennial, biennial, and winter annual weeds harboring CMV in roots, tubers and underground organs throughout the winter include common milkweed (Asclepias syriaca), yellow rocket (Barbarea vulgaris), marsh yellowcress (Rorippa islandica), and yellow toadflax (also called butter-and-eggs, Linaria vulgaris). These were shown to be important sources for infection of lettuce in upstate New York. Seven additional weeds, including common chickweed (Stellaria media) were shown to be important overwintering sources in Britain for lettuce. It is important to note that infected weeds are often symptomless.
Crop hosts may also serve as significant sources of CMV for other crops. A good example of this is how the perennial crop alfalfa can serve as the reservoir of CMV for subsequent infection of snap beans. Although alfalfa is symptomless, it is a formidable reservoir of CMV, and is currently a major production problem for snap beans grown in Wisconsin, Pennsylvania and New York.
Seed as a source of CMV has been reported in common chickweed and in 19 other plant species. In chickweed (Caryophyllaceae), the rate of transmission was as high as 40% in plants grown from infected seed. Other plant families with seedborne CMV (including crop plants) are Amaranthaceae (syn. Chenopodiaceae) (spinach), Brassicaceae, Fabaceae (syn. Leguminosae) (bean, chickpea, cowpea, lentil, lupin, subterranean clover) and Labiatae. This seed transmission feature has proven to be important for the annual occurrence of CMV-infected legumes in commercial fields, and for the potential infection of greenhouse crops growing close to chickweed plants surviving in protected areas near greenhouses. The seedborne characteristic increases the probability that the virus will survive in nature.
More than 80 aphid species (Insecta: Hemiptera: Aphidoidea), including Myzus persicae and Aphis gossypii, are capable of transmitting the virus in a nonpersistent, stylet-borne manner (Figure 10). The nonpersistent relationship implies that the insect acquires the virus in short probes (generally less than 60 sec.) and retains the virus for short periods of time (a few minutes) and loses the virus due to normal feeding activities. This means that the insect would have to reacquire the virus again (become viruliferous) to be able to transmit again. In the case of snap bean, the most important aphid vectors are the soybean aphid (Aphis glycines, recently introduced into the US), the yellow clover aphid (Therioaphis trifolii) and less importantly, the pea aphid (Acyrthosiphon pisum). The virus is rapidly acquired by all instars, generally within one minute of feeding, but their ability to transmit the virus declines and is lost within several hours. Transmission efficiency varies with the aphid species, virus strains, host plant species, environmental conditions and time of the year. Experimentally, CMV is easily transmitted mechanically, but efficiency is increased by addition of a reducing agent such as 10mM sodium sulfite to the buffer when preparing inoculum. Experimental mechanical transmission involves grinding infected tissue in a buffer (typically in a mortar with a pestle); many viruses can remain infectious during this process by using a simple buffer to resist pH change and protect the virus particle. Some viruses, like CMV and Tomato spotted wilt virus, are less stable and more vulnerable to being degraded during this process. Addition of a reducing agent can help to stabilize the virus, thereby enhancing its ability to remain infectious for transmission. Since CMV is not a stable virus like Tobacco mosaic virus, it is not likely to be transmitted by workers touching infected plants during normal field operations.

Figure 10. Winged green peach aphid. Click the image to enlarge, or click here for a sample video clip of non-persistent transmission of a virus (the video shows PVY, not CMV).
View Video
|
Disease Management
Development of genetic resistance is the simplest and most effective method of controlling virus diseases and is especially appropriate for CMV. Success has been noted, especially for cucumber and spinach, but is variable among most other crops. The history of CMV resistance in cucumber (Cucumis sativus) dates back to 1927 when the oriental varieties ‘Chinese Long’ and ‘Tokyo Long Green’ were introduced to the US. After extensive study, it was concluded that homozygosity of three partially dominant genes was needed to convey a high level of resistance for cucumber. This resistance formed the background for all modern day slicing and pickling cucumber varieties and has been effective for many decades, perhaps because resistance is based upon several genes.
Progress in finding resistance in other cucurbit species has not been as successful. Even though resistance has been found in other Cucurbit species, genetic incompatibility among species remains a formidable barrier for transferring this resistance to Cucurbita pepo crops, such as summer squash. The precocious gene, which masks color breaking in yellow summer squash for CMV (and WMV), is present in many yellow straightneck and crookneck varieties, and several have transgenic resistance or intermediate resistance for CMV. Insertion of the precocious gene is by traditional cross breeding whereas transgenic resistance involves the insertion of one or more foreign genes from a different organism. A handful of zucchini squash varieties have intermediate resistance for CMV and a few have transgenic resistance.
Genetic resistance for CMV in melon (Cucumis melo) is derived from oriental melons, and depending on the source, resistance is controlled by two or three complementary recessive genes. Some of these factors for resistance are strain and/or temperature dependent, with plants developing symptoms at temperatures below 20°C. No commercial muskmelon varieties with CMV resistance are available. Fortunately, most varieties of watermelon (Citrullus lanatus) are resistant to the most prevalent strains of CMV, with the exception of a specific strain that can infect plants systemically.
CMV resistance in spinach is controlled by a single dominant gene in the variety Virginia Savoy, and this has been incorporated into many current spinach varieties. This resistance, however, is not complete and may break down at temperatures greater than 28°C.
Virus resistance in lettuce to CMV was identified in Lactuca saligna PI 26153 from Portugal, a distantly related species of lettuce (L. sativa). However, this resistance is strain specific. An accession from L. serriola proved to be tolerant to three strains of CMV studied, but this tolerance has not been transferred to any current commercial variety.
Extensive effort was deployed to develop pepper resistance for CMV, and although tolerance has been described in some varieties, such as ‘Perennial’, it was not utilized in any commercial variety. More recently the pepper variety ‘Peacework’ was developed with a high level of resistance.
Sources of resistance and tolerance to CMV have been reported in wild tomato relatives but not in Solanum lycopersicum. No tomato varieties with CMV resistance have been released.
Eradication of weed hosts is often a difficult task because of the extensive host range of this virus. However, elimination of several of the key perennial or biennial weeds located near the crop may reduce severe virus pressure, and has successfully been used to control CMV.
In addition, isolation of crop fields by growing taller, nonsusceptible barrier crops such as corn, may delay initial infections. Application of insecticides and mineral oil sprays has been used to control CMV, as well as other nonpersistently transmitted viruses, but mineral oil sprays are most effective for crops with high plant populations per acre (e.g. pepper and tomato) and less so for crops with low plant populations (e.g. vining crops like cantaloupes or watermelons). The easiest explanation for this phenomenon is that when using a protective spray like mineral oil, and trying to delay infection for a matter of weeks, the probability of obtaining an infection in a low acreage crop is much greater than in a crop with higher plant populations. Insecticide applications are very effective for aphid control, but do not act quickly enough to prevent insect transmission because of the nonpersistent manner of transmission.
One method that has been used successfully to prevent infection of a susceptible crop like cantaloupe has been to use a Remay floating row cover to exclude migrant aphids during the early weeks of crop growth. Crops that are “protected” during the main period of aphid migration and transmission can remain free of virus infection, often to the end of the season. The covers are removed after this period of greatest vulnerability has passed so that bees can successfully pollinate the crop.
Alternative methods of reducing CMV losses have also been reported. Cross-protection, the inoculation of plants with mild, protective strains of CMV or the use of satRNAs (see Pathogen Biology), have been described in China. Transgenic tomato genotypes, expressing either the CMV coat protein or a satRNA, protected plants from CMV infection or the severe effects of some satellite RNA-associated CMV infections. The application of resistance-inducing bacterial treatments that may also enhanced plant growth provided protection against CMV, and may be integrated into other pest management schemes. There are numerous commercial products available that consist of single, or more often, multiple species of bacteria that colonize plant roots or root zones. These bacteria induce or enhance a plant’s ability to resist infection and, in some cases, have been effective against CMV.
Significance
Cucumber mosaic virus occurs worldwide and is considered a very important disease in temperate, tropic and subtropic regions of the world. Crop losses vary from year to year since the amount of disease occurrence depends upon the number of aphids available for virus transmission in the spring or fall when the crops are established as determined by geographical location. If the spring or fall is cool and wet, aphid numbers are decreased and virus spread is sporadic, with infected plants primarily located in rows bordering the edges of the field. However, if the spring or fall is warmer with less frequent rains, aphid populations increase rapidly on perennial crops that harbor CMV, and virus spreads rapidly into crops that are young and especially attractive to migrating aphids. In such cases, infection rates may approach 100% and the crop may have to be abandoned. On average, losses of 10-20% are common, and in some instances the crop may still be harvested, but is of poorer quality and appearance.
The virus can infect nearly 1,200 plant species. The host range includes many important vegetable crops including artichoke, beans (broad, lima, snap) [Figure 4 I], beet (garden and sugar), carrot, celery [Figure 4 F, G], cucurbits (chayote, cucumber, loofah, melon, pumpkin, summer and winter squash, watermelon) [Figure 3 A-E], lettuce [Figure 4 E], pea, pepper [Figure 4 A-C], potato, sweet potato (Ipomoea), spinach [Figure 4 D], and tomato [Figure 4 H, 5, 6]). Other crops affected include alfalfa (symptomless), banana (symptomless or mild to severe damage), chickpea, lupin, oil seed rape, subterranean clover, and yams (Dioscorea spp.). Ornamentals infected include anemone, candytuft, columbine, dahlia, delphinium, geranium, gladiolus, lily, marigold, petunia, phlox, viola, zinnia, etc., and woody and semiwoody plants (ixora, passion fruit, etc.). Numerous isolates of CMV have been reported as strains, some of which are specific for given crops.
Selected References
Abad, J., G. Anastasio, A. Fraile, and F. García-Arenal 2000. A search for resistance to Cucumber mosaic virus in the genus Lycopersicon. Journal of Plant Pathology 82:39-48.
Doolittle, S.P. 1920. The mosaic disease of cucurbits. United States Department of Agriculture Bulletin 879. 69 pp.
Francki, R.I.B., D.W. Mossop, and T. Hatta, T. 1979. Cucumber mosaic virus. Descriptions of Plant Viruses, No. 213 (No. 1 revised). Commonwealth Mycological Institute, Association of Applied Biologists, Kew, Surrey, England.
Gallitelli, D. 2000. The ecology of Cucumber mosaic virus and sustainable agriculture. Virus Research 71:9-21.
Gonsalves, D., R. Provvidenti, and M.C. Edwards. 1982. Tomato white leaf: the relation of an apparent satellite RNA and cucumber mosaic virus. Phytopathology 72:1533-1538.
Hooks, C.R.R., and A. Fereres. 2006. Protecting crops from non-persistently aphid-transmitted viruses: a review on the use of barrier plants as a management tool. Virus Research 120:1-16.
ICTVdB Management. 2006. 00.010.0.04.001. Cucumber mosaic virus. In: ICTVdB - The Universal Virus Database, version 4. Büchen-Osmond, C. (Ed), Columbia University, New York, USA. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/00.010.0.04.001.htm
Jones, R.A.C., B.A. Coutts, L.J. Latham, and S.J. McKirdy. 2008. Cucumber mosaic virus infection of chickpea stands: temporal and spatial patterns of spread and yield-limiting potential. Plant Pathology 57:842-853.
Kyle, M.M. (ed.) 1993. Resistance to viral diseases of vegetables, genetics and breeding. Timber Press, Portland, OR.
Mazourek, M., G. Moriarty, M. Glos, E. Henderson, D. Rumore, G. Palmer, A. Chickering, J.F. Murphy, M. Fink, M. Kreitinger, C. Kramer, D. Kean, J.R. Myers, and M. Jahn. 2009. ‘Peacework’: A cucumber mosaic virus resistant early red bell pepper for organic systems. HortScience. In press.
Murphy, J.F., E.J. Sikora, B. Sammons, and W.K. Kaniewski. 1998. Performance of transgenic tomatoes expressing cucumber mosaic virus CP gene under epidemic conditions. HortScience 33:1032-1035.
Murphy, J.F., M.S. Reddy, C.–M. Ryu, J.W. Kloepper, and R. Li. 2003 Rhizobacteria-mediated growth promotion of tomato leads to protection against Cucumber mosaic virus. Phytopathology 93:1301-1307.
Palukaitis, P., M.J. Roossinck, R.G. Dietzgen, and R.I.B. Francki. 1992. Cucumber mosaic virus. Advances in Virus Research 41:281-348.
Palukaitis, P., and F. García-Arenal. 2003. Cucumoviruses. Advances in Virus Research. 62:241-323.
Rist, D.L., and J.W. Lorbeer. 1989. Occurrence and overwintering of cucumber mosaic virus and broad bean wilt virus in weeds growing near commercial lettuce fields in New York. Phytopathology 79:65-69.
Simons, J.N. and T.A. Zitter. 1980. Use of oils to control aphid-borne viruses. Plant Disease 64:542-546.
Tomlinson, J.A., and A.L. Carter. 1970. Studies on the seed transmission of cucumber mosaic virus in chickweed (Stellaria media) in relation to the ecology of the virus. Annals of Applied Biology 66:381-386.
