Papaya ringspot
Papaya ringspot virus
Cucurbits and papaya
Symptoms and Signs
Papaya ringspot virus infects papaya and cucurbits systemically. Symptoms on papaya are somewhat similar to those on cucurbits. In papaya, leaves develop prominent mosaic and chlorosis on the leaf lamina, and water soaked oily streaks on the petioles and upper part of the trunk. Severe symptoms often include a distortion of young leaves which also result in the development of a shoestring appearance that resembles mite damage. Trees that are infected at a young stage remain stunted and will not produce an economical crop. Fruit from infected trees may have bumps similar to that observed on fruit of plants with boron deficiency and often have ‘ringspots’, which is the basis for the disease’s common name (Figure 1). A severe PRSV isolate from Taiwan is also known to induce systemic necrosis and wilting along with mosaic and chlorosis.

Figure 1
In cucurbits, leaves show intense mosaic with a narrowing of the leaves. Severe cases can result in a shoestring effect similar to what is observed in papaya. Plants that become infected at a young age do not develop a crop. Older plants that become infected do produce fruit that often show distinct changes in color and are deformed. For example, a yellow crook neck squash would have many green blotches and bumps (Figure 2).

Figure 2
The experienced eye can detect symptoms on infected papaya at a very early stage before intense mosaic symptoms and chlorosis develop. The first symptoms are the appearance of oily streaks on the younger leaves and the younger leaves show clearing along the veins that gives an appearance of flecks. These early symptoms are used to detect infected plants when rogueing is used for managing the disease.
Pathogen Biology
The general properties of PRSV are similar to the vast majority of viruses in the genus Potyvirus of the family Potyviridae. Aphids transmit the virus to papaya and cucurbits in a nonpersistent manner; in other words, the virus is acquired and transmitted by its vector in short periods of time that are measured in seconds to a minute. The virus does not replicate in the vector. The amorphous inclusion (AI) protein, a helper component protein which is a product of the virus gene (HC-Pro), is required for successful vector transmission. In addition to aphids, PRSV is also easily transmitted via mechanical inoculation but there are no confirmed reports of PRSV transmission through seeds.
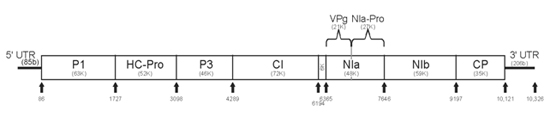
The virus particle or virion consists of a nucleocapsid, a filamentous flexuous rod measuring roughly 760-800 x 12 nm (Figure 3). Virus particles typically contain 94.5% protein and 5.5% nucleic acid by weight, and have no outer membrane (non-enveloped). The buoyant density of the purified virions is 1.32 g cm-3 in cesium chloride. The thermal inactivation point (TIP) of PRSV is 54-60°C and the longevity in vitro (LIV) is about 0.3 days. The first complete PRSV genome sequence was obtained from a PRSV isolate from Hawaii (PRSV HA). Like other potyviruses, PRSV has a monopartite linear single-stranded positive sense RNA genome and is about 10,326 nucleotides long, excluding a poly-A-tract found at its 3’ end. Typical of potyviruses, the PRSV genome encodes a single large protein, (in the case of PRSV, 3,344 amino acids) which is subsequently cleaved into smaller proteins with various functions. The cleaved proteins are: P1, HC-Pro, P3, CI, 6K, NIa-Pro, NIb and CP (Figure 4). The different functional proteins are formed by a series of site-specific cleavage events carried out by three virus-encoded proteases, P1, HC-Pro, and NIa. The possible functions of the PRSV genome encoded proteins have been deduced from various potyvirus studies and are listed in Figure 4.
Figure 3 |
|
Genes |
Size (Mr) |
Functions |
|
P1 |
63K |
Proteinase |
|
|
|
Cell-to-cell movement |
|
HC-Pro |
52K |
Vector transmission |
|
|
|
Proteinase |
|
|
|
Pathogenicity |
|
|
|
Suppressor of RNA silencing |
|
|
|
Cell-to-cell movement |
|
P3 |
46K |
Unknown, but possible role in replication |
|
6K1 |
6K |
Unknown, but possible roles in: |
|
|
|
RNA replication |
|
|
|
Regulation; inhibition of NIa nuclear translocation |
|
|
|
Replication |
|
CI |
72K |
Genome replication (RNA helicase) |
|
|
|
Membrane attachment |
|
|
|
Nucleic acid stimulated ATPase activity |
|
|
|
Cell-to-cell movement |
|
6K2 |
6K |
Same as 6K1 |
|
N | Ia-VPg |
21K |
Genome replication (Primer for initiation of RNA synthesis) |
|
NIa-Pro |
27K |
Major Proteinase |
|
NIb |
59K |
Genome replication (RNA-dependent RNA polymerase, RdRp) |
|
CP |
35K |
RNA encapsidation |
|
|
|
Vector transmission |
|
|
|
Pathogenicity |
|
|
|
Cell-to-cell movement |
Figure 4. PRSV genome organization, genes and their possible functions. (Courtesy of D. Gonsalves, copyright-free)
Interestingly, PRSV is divided into two major biotypes or strains based on their host range. The PRSV-W type affects cucurbits but not papaya, while the PRSV-P type affects papaya in addition to cucurbits. This grouping was done to clarify historical literature as well as to provide an indication of the life cycle and impact on the epidemiology of the disease.
Prior to 1984, when the P and W biotypes were separated, the literature had labeled the disease of papaya caused by what we know today as PRSV-P, papaya ringspot and early reports showed that the virus affected papaya and also affected cucurbits. In parallel, a viral disease of cucurbits caused by what we now know as PRSV-W was designated as Watermelon mosaic virus-1 (WMV-1) to distinguish it from another disease that was caused by Watermelon mosaic virus-2 (WMV-2). There was not an obvious connection between PRSV in papaya and WMV-1 in cucurbits since WMV-1 did not affect papaya. Later serological studies, however, showed that PRSV and WMV-1 were serologically indistinguishable and thus it was suggested that they were biotypes of the same virus, Papaya ringspot virus. This functional grouping has served the scientific community well because it provides a clear and simple way to distinguish the biotypes.
The PRSV, virus code:00.057.0.01.045, is an ICTV approved species of the genus Potyvirus (00.057.0.01) in the family Potyviridae (00.057). A useful link for this virus is: ICTVdB Index of Viruses.
Disease Cycle and Epidemiology
The virus, PRSV, is transmitted nonpersistently by aphid vectors and does not multiply in the vector. The disease cycle can start with aphids feeding on infected papaya for as little as 15 seconds and subsequently feeding on a healthy papaya. There is no incubation period. The virus does not persist in the vector so transmission to another plant has to occur rather rapidly. For a brief video clip of this type of transmission, see Video clip - Cucumber mosaic virus. The PRSV-P and W biotypes are found wherever their host crops are grown. For example, the W biotype is found extensively in areas where there is little papaya grown, such as in the mainland U.S. In fact, PRSV-W is one of four major viruses (PRSV-W, WMV-2, Zucchini yellow mosaic virus, and Cucumber mosaic virus) that cause chronic damage to cucurbits in the southeastern U.S. However, PRSV-W is also found in and causes damage to cucurbits in tropical and subtropical regions. On the other hand, PRSV-P is only found in tropical and subtropical areas where papayas are normally grown.
In the tropics and subtropics where both biotypes and their hosts are present, PRSV-P effectively completes its life cycle in papaya while PRSV-W completes its life cycle in cucurbits. In other words, cucurbits generally do not serve as alternative hosts for PRSV-P.
Observations on PRSV in Australia have provided some insights as to the origin of the PRSV biotypes. Until 1991, only PRSV-W had been observed in Australia. However, in 1991, PRSV-P was discovered there in papaya. Sequence analyses of the ‘new’ PRSV-P as well as the PRSV-W type in Australia showed that they were very closely related.
Thus, it was surmised that the appearance of PRSV-P in Australia was due to mutations in the established PRSV-W that provided the ability to infect papaya. These observations suggest that PRSV-P originated from PRSV-W. In fact, it has been shown experimentally, that only a few point mutations distinguish the PRSV-P and PRSV-W biotypes.
Disease Management
The various options for managing viral diseases are vector control, planting in areas where there is no virus, rogueing, cross protection, and resistance. Vector control for managing PRSV is not an economical option for several reasons. PRSV is nonpersistently transmitted and thus the vector acquires and transmits the virus within seconds to minutes. Thus, aphids coming into target areas of vector control could acquire and shortly thereafter transmit the virus, and insecticides cannot kill them fast enough. As mentioned above, the economically relevant host range of PRSV includes papaya and cucurbits, two crops that are very different in growth characteristics. Papaya is a large perennial tropical herbaceous tree while cucurbits are vegetable crops that are grown over a period of only several months.
Papaya. Papaya is not a preferred host of the PRSV vectors, aphids. Disease management by controlling the vector within the papaya orchard is very difficult as aphids visit and probe papayas but do not colonize papaya orchards.
Classical resistance and tolerance in papaya. Natural resistance to PRS in Carica papaya has not been identified. Resistance to PRSV exists in Vasconcellea species, which are related to Carica papaya. However, crosses between Carica papaya and Vasconcellea species are not fertile. Efforts to transfer the putative resistance gene(s) by crossing of Carica papaya and Vasconcellea species followed by embryo rescue and subsequent back crossing with Carica papaya has been attempted, but so far have not been developed to the stage of commercial cultivation. Efforts to incorporate tolerance to PRS in Carica papaya have been a bit more successful. Some PRS tolerant lines have been commercialized in the Philippines and are sold commercially in Taiwan, and tolerant lines have been released in Florida and Thailand; these lines develop only mild symptoms when infected. PRSV can still spread throughout the orchards of tolerant lines, since they are not resistant to the virus. When these tolerant lines do become infected with PRSV, they are still able to produce respectable crops. However, the quality of fruit from these infected tolerant lines is generally marginal which leads to their not being widely grown.
Cross protection. Cross protection is the phenomenon whereby plants that are systemically infected with a mild strain of a virus are protected against the effects of infection by a more virulent related strain. Cross protection was practiced quite extensively in Taiwan and to a very limited extent in Hawaii both in the 1980s. At both locations, the same mild strain was used, which was developed by selecting a mild mutant from nitrous acid treatment of tissue extracts of leaves infected with a severe strain of PRSV from Hawaii. This mutant strain caused mild symptoms on both papaya and cucurbits and gave good cross protection against severe strains of PRSV in Hawaii. However, the first attempts to use this mild strain for large scale protection was initiated in Taiwan, where PRSV was widespread and causing severe damage. The general steps followed for cross protection of papaya were: production of inoculum, which consisted of infection of Cucumis metuliferus (horned melon) with the mild PRSV strain, spray inoculation of young papaya seedlings with extracts from infected C. metuliferus plants in the greenhouse or net house, and planting the seedlings in the field 3-4 weeks after establishment of the virus. These plants would subsequently be protected against damage caused by severe strains of the virus. Cross protection was practiced on a large scale in Taiwan for several years, but the practice was abandoned for several reasons. The logistics of producing the mild strain inoculum and subsequently ensuring the supply of mild strain inoculated papaya seedlings added a significant step in the production of papaya. This added step would have been worthwhile, but results from the large scale application in Taiwan showed that the mild strain did not protect as well against the Taiwanese PRSV strains as it did against the virulent Hawaiian strain. After awhile farmers were not enthusiastic to continue the management practice of cross protection in Taiwan.
In Hawaii, cross protection worked very well in limited field trials on the island of Oahu where PRS was very severe. Even in Hawaii, however, the practice was not fully commercialized because the mild strain caused quite severe symptoms on some cultivars such as Sunrise and because the logistics for raising the inoculum, uniformly infecting plants with the mild strain and delivery of the mild strain-infected plants to the farmers did not work out economically.
Genetically engineered resistance in papaya. Genetically engineered (GE) papaya has been produced and commercialized in Hawaii to successfully control PRS since 1998. This is the first fruit crop to be genetically engineered and commercialized. What follows is some background to the story of the development of PRS-resistant GE papaya in Hawaii.
The state of Hawaii includes seven inhabited islands (Hawaii, Maui, Oahu, Kauai, Molokai, Lanai, and Niihau) isolated by miles of ocean. It is, thus, buffered from continual influx of PRSV from other geographic regions. PRSV was discovered on the island of Oahu in the 1940s where Hawaii’s papaya industry was located. In the 1950s, PRSV caused severe damage to papaya orchards on Oahu, and thus papaya production was relocated to the Puna district on the island of Hawaii in the early 1960s. In Puna, the industry enjoyed many years of papaya production in the absence of PRSV. However, PRSV was detected and became established in Hilo on the island of Hawaii in the late 1960s. Other factors in addition to geographic isolation played key roles in keeping PRSV out of Puna since Hilo was only 19 miles away. The other factors included continually surveying and rogueing of PRS-affected papaya in Hilo, activities that helped to lower the incidence of PRSV in the Hilo area, a quarantine on the movement of papaya seedlings into Puna, and surveillance for PRS in the Puna district where by the 1970s, 95% of the state of Hawaii’s papaya were being produced. Thus, a combination of avoidance, geographical isolation, rogueing, use of quarantine to keep infected seedlings from being transported to Puna, and constant efforts to detect and eliminate infected trees all contributed to keeping the Puna district free of PRSV.
However in 1992, PRSV was detected in Puna and the scenario that developed in the ensuing 5 years provides a realistic assessment on management of PRS in plantations where there is little chance of complete avoidance. The situation was described previously in 1998 and is quoted here since it provides the sense of urgency that the Hawaii papaya industry faced during 1992 to 1997 as is also evident by photos taken of the area during the same period (Figure 5).

Figure 5
Quote from the Annual Review of Phytopathology:
The inevitable entry of PRSV into the Puna district on Hawaii island was discovered during the first week of May in a papaya field in Pahoa, 1-3 miles from the major papaya growing areas in Puna (23). Apparently, infection had been established in this area for several months, as judged by symptoms on the fruits and the fact that many plants were infected in one location. Surveys of the immediate area revealed PRSV in abandoned orchards, as well as in young orchards that were not yet producing fruit. PRSV was poised to invade the major papaya growing areas of Puna, which included Kapoho, Opihikau, Kahuawai, and Kalapana.
The Hawaii Department of Agriculture (HDOA) immediately launched an eradication program. The area was surveyed and infected trees were rouged [sic] out. A suggestion to destroy all papaya in that area, however, was not approved by the growers. Nevertheless, a HDOA program to mark trees to be rouged by growers was started in 1992 (23). By September 1992, 4915 trees had been rouged in Pahoa, and the number of trees being cut each week had decreased to below 85, providing hope that the virus had been contained (23).
However, the hope of containment was short-lived. The incidence of PRSV increased dramatically in Kapoho, which was closest to Pahoa, as the program of voluntary cutting of trees was not strictly followed and as farmers experiencing high infection rates abandoned their fields, thus creating huge reservoirs of inoculum for aphids to acquire and spread the virus (24). By late 1994, nearly all papaya of Kapoho was infected by the virus. In October 1994, the HDOA declared that PRSV was uncontrollable and stopped the practice of marking trees for rouging. In less than three years, a third of the Puna papaya area was infected… … Five years after the onset of the virus in Pahoa, the entire Puna area was severely affected.
Investigations to develop GE papaya with resistance to PRS began in 1985. The concept of pathogen-derived resistance or parasite-derived resistance was employed. This concept basically states that plants that contain gene(s) or sequences of a pathogen are protected against detrimental effects of the same or closely related pathogens. For papaya, our approach was to develop transgenic papaya with the coat protein (CP) gene of PRSV which would protect the transgenic plant against damage caused by PRSV. Briefly, the research to obtain the transgenic papaya involved isolating and sequencing the CP gene of PRSV from Hawaii, transforming embryogenic calli of nontransgenic Sunset papaya, a commercial papaya in Hawaii, selecting and regenerating plants transformed with the CP gene of PRSV, screening transformed plants for resistance to PRSV, field trials, deregulation, and commercialization.
In 1991, a PRS-resistant GE papaya line designated 55-1, was identified (Figure 6) and subsequently field tested for resistance (Figure 7). Papaya line 55-1 was used to create the cultivars SunUp and Rainbow. SunUp is a transgenic version of its red-fleshed, progenitor cultivar Sunset and is homozygous for the CP gene. Rainbow is an F1 hybrid created by crossing transgenic SunUp with nontransgenic Kapoho. The latter was the dominant cultivar growing in Puna when PRSV was discovered there in 1992. The transgenic cultivars were field tested, deregulated by the U.S. government, and subsequently commercialized upon release of seeds of SunUp and Rainbow to growers in 1998.

Figure 6 |

Figure 7 |
The transgenic papaya, especially Rainbow, allowed the papaya farmers to reclaim Puna from the ravages of PRS (Figure 8). Use of genetic engineering allowed introduction of resistance properties otherwise not available in existing germplasm, directly into commercially viable cultivars without altering other traits. The engineered trait could be stably transmitted genetically by seed. This allowed the rapid development of cultivars that saved the Hawaiian papaya industry and that currently represent over 70% of the planted papaya acreage. Stable genetic transmission of the 55-1 line transgene also allowed the development of new niche market papaya cultivars with the same resistance properties.

Figure 8
Cucurbits. PRSV-W along with other potyviruses including Zucchini yellow mosaic virus (ZYMV) and WMV is a major limiting factor in cucurbit production worldwide. Aphids in fact colonize cucurbits more than papaya and vector control for managing PRSV-W in cucurbits has not been successful with PRSV nor for most potyviruses that infect cucurbits. Some success has been demonstrated in the northeastern U.S. particularly with summer squash and melon by using cover crops in the spring to prevent the early influx of viruliferous aphids. Reflective mulches that work to repel aphids by reflecting UV light are also useful, but mainly in desert areas with regular sunshine. However, general cultural practices for managing viral diseases are unable to satisfactorily control PRSV in commercial cucurbit crops.
Unlike papaya, genetic resistance to PRSV has been identified in cucurbit species and these have been successfully incorporated into commercial cultivars of cucurbits through various breeding programs. A list of commercial cultivars resistant/tolerant to PRSV-W can be found at: Vegetable MD Online.
The incorporation of resistance/tolerance in cucurbits by breeding has been successful and widely used for the production of popular cucurbit cultivars. The planting of virus-resistant or tolerant cultivars also greatly reduces virus pressure and subsequently the chances of developing a virus epidemic even for susceptible crops grown in the same vicinity.
Resistance to PRSV-W is controlled by a single dominant gene in cucumber and a single dominant gene in melon. In squash, resistance to PRSV-W is controlled by a single recessive gene identified in C. moschata and by three partially-dominant genes in C. maxima. Sources of resistance to PRSV-W have also been reported in watermelon. Extensive screening for resistance to PRSV-W in the USDA watermelon germplasm collection (total of 1650 accessions) identified a single recessive locus (prv) in C. lanatus var. citroides plant introduction accessions (PI) from South Africa (PI 244017, PI 244018, PI 244019), Zimbabwe (PI 482342, PI 482318, PI 482379), and Botswana (PI 485583). Resistance was also identified in a C. lanatus var. lanatus accession from Nigeria (PI 595203). All virus resistance data on PI accessions have been submitted to The Germplasm Resources Information Network (GRIN) for future use by researchers.
As cucurbits are often infected by other potyviruses simultaneously, resistance to PRSV-W alone is insufficient for commercial production. The development of new cultivars with resistance to multiple viruses by conventional breeding can be difficult, costly and time consuming because, along with resistance genes, desired horticultural traits must also be selected for.
Genetic engineering using the approach of pathogen-derived resistance is one of the most effective and efficient means to develop varieties with resistance to several viruses. In fact, several squash varieties with resistance to multiple viruses that commonly infect cucurbits have been developed by GE techniques. If natural resistance exists, combining conventional breeding with GE approaches can be useful and efficient for developing new cultivars with broad spectrum resistance. A good example is green stem straightneck summer squash cultivar Conqueror III developed by Seminis. Conqueror III has transgene-derived resistance to the potyviruses, ZYMV and WMV, the cucumovirus, Cucumber mosaic virus (CMV) as well as conventionally bred natural tolerance to PRSV-W.
Significance
PRS is one of the most destructive diseases of papaya and occurs in nearly every region where papaya is grown. It has been reported to be a major limiting factor for commercial papaya production particularly in Hawaii, areas of Thailand, Taiwan, India, Mexico, Brazil, Bangladesh, the Philippines, and the southern region of China. In addition, PRSV-W is also a cause of serious losses worldwide in several important cucurbit crops of tropical, subtropical and temperate regions.
PRS has had historical, economical, scientific and cultural significance for the Hawaiian papaya industry for more than 70 years since the time it was first discovered in the 1940s on the island of Oahu. Initially it caused relatively mild symptoms until the mid 1950s when a new severe PRSV strain initially described as Papaya mosaic virus emerged either from the original milder strain of the virus or as a new introduction. Within just 10 years of its occurrence, the new yellow mosaic strain of PRSV became widespread and was able to severely impact the papaya industry as described previously.
Development of PRS resistant transgenic papaya cultivars Rainbow and SunUp and their timely commercial release in Hawaii in 1998, during the time of disease crisis played a key role in saving and reviving the papaya industry in Hawaii. Currently, transgenic papaya is the dominant cultivar grown in Hawaii and as of 2010 accounts for more than 70% of Hawaii’s papaya acreage. Rainbow and SunUp have been commercially grown and consumed in Hawaii and the rest of the U.S. for more than a decade without any reports of adverse effects on human health.
Hawaii’s transgenic papaya was also deregulated in Canada and is soon expected to be approved in Japan. The amassed safety data has made GE papaya (line 55-1) one of the most well characterized transgenic plants, providing significant contributions to science and education in this area. Transgenic papaya approval in Japan would not only have great impact to Hawaii’s papaya industry, but would also advance the case of transgenic products outside of the U.S. as it will be the first GE fresh fruit crop commercialized there. If approved, consumers in Japan will have the opportunity to directly compare between a GE and non-GE fresh product. Marketing of GE papaya in Japan will educate us on the factors influencing consumer acceptance of fresh GE products outside of the U.S. in a real life scenario.
Selected References
Gonsalves, D., and M. Ishii. 1980. Purification and Serology of Papaya Ringspot Virus. Phytopathology 70:1028-1032
Gonsalves, D. 1998. Control of papaya ringspot virus in papaya: A case study. Annual Review of Phytopathology 36:415-437.
Gonsalves, D., C. Gonsalves, S. Ferreira, K. Pitz, M. Fitch, R. Manshardt, and J. Slightom. 2004. Transgenic virus resistant papaya: From hope to reality for controlling papaya ringspot virus in Hawaii. APSnet feature story for July, 2004.
Gonsalves, C., D. R. Lee, and D. Gonsalves. 2004. Transgenic virus-resistant papaya: The Hawaiian 'Rainbow' was rapidly adopted by farmers and is of major importance in Hawaii today. APSnet feature story for August-September 2004.
Gonsalves, D., J. Y. Suzuki, S. Tripathi, and S.A. Ferreira. 2007. Papaya ringspot virus (Potyviridae). In: B.W.J. Mahy and M.H.V. van Regenmortel (Eds.), Encyclopedia of Virology, 5 vols. Elsevier Ltd, Oxford, UK.
Purcifull, D., J. Edwardson, E. Hiebert, and D. Gonsalves. 1984. Papaya ringspot virus. CMI/AAB Descriptions of Plant Viruses. No. 292. (No. 84 Revised, July 1984). 8 pp. CAB International, Wallingford, UK
Tripathi, S., J. Y. Suzuki, S.A. Ferreira, and D. Gonsalves. 2008. Papaya ringspot virus-P: characteristics, pathogenicity, sequence variability and control. Molecular Plant Pathology 9:269-280.
Yeh, S. D., F. J. Jan, C. H. Chiang, P. J. Doong, M. C. Chen, P. H. Chung, and H.J. Bau. 1992. Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. Journal of General Virology 73:2531-2541.
