Basic concepts in Bayesian analysis
Bayesian analysis is the statistical methodology for updating existing knowledge with new information. One begins with a prior belief about a parameter and updates the belief after obtaining new information from data (e.g., from an experiment or a survey). Here, a parameter loosely denotes some quantity of interest, such as the mean (expected value) of a distribution (e.g., length of the spores of a pathogen species) or slope of a line (e.g., disease increase over time). In classical (frequentist) statistics, parameters estimated in data analysis are constants, but in Bayesian analysis, estimated parameters are random variables that are characterized by distributions.
With Bayesian analysis, all information is expressed in terms of probability distributions and there are three distributions that are critically important: (i) the distribution of the observations (e.g., normal, Poisson, binomial distribution); (ii) the prior distribution of the parameters; and (iii) the posterior distribution of the parameters. The ultimate goal is to estimate the posterior distribution of the parameters using the first two distributions. Before discussing the specifics, it is important to review some concepts related to random variables and probability distribution.
Elements of Probability
Why is probability important?
Here we will outline a few reasons that are most relevant to Bayesian analysis, as to the importance of probability.
(i) Any process with an uncertain outcome is a
random process. If the random process can be described numerically, then it is known as a random variable.
Probabilities are excellent descriptors of random processes (or variables). A process can be either (i) Discrete (when outcomes can be put in a list of separate items) or (ii) Continuous (when outcomes can take any value in a sample space).
Let’s assume, for example, a binomial process that can be described by the expected value (or mean) and the variance:
The
expected value E(y) of a
binomial random variable can be computed using the simple analytic expression: E(y) =
np. Here,
y represents the random variable (a count from 0 to
n),
n is the number of observations (trials, etc.), and
p is the probability of success.
The variance of a binomial random variable can be computed using the analytic expression: V(y) =
np (1-p) =
n×p×q, where
q = (1-p) by definition.
(ii) Probabilities describe uncertainty in a quantitative manner. For instance if we use the distributions in Fig. 1, we can conclude for the distribution on the left that “90% of the random variable X is between the values of 5 and 10” or for the distribution on the right that “the probability the random variable Y is above 7 is 0”. These are only examples but several more statements can be made depending on the question that is being asked.
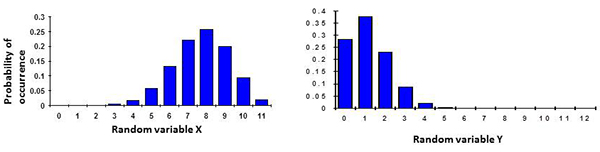
Figure 1. (left)
If
p is large, the distribution tends to be skewed with a tail on the left (p = 0.7 in example); and (right) if
p is small, the distribution tends to be skewed with a tail on the right (p = 0.02 in example).
Click to enlarge.
(iii) Basic rules of probability allow algebraic manipulations to express the probability of interest in terms of other probabilities. Suppose
y and
 are random variables and P(•) represents probability, either as a point value (e.g., 0.5) or a probability distribution (e.g., normal distribution). We use
y for the random variable representing the data collected in an experiment or a survey, and
are random variables and P(•) represents probability, either as a point value (e.g., 0.5) or a probability distribution (e.g., normal distribution). We use
y for the random variable representing the data collected in an experiment or a survey, and  for a parameter of interest. Bayesians consider
for a parameter of interest. Bayesians consider  as a random variable because there is uncertainty in its value, before and after data are collected and analyzed. P(y|
as a random variable because there is uncertainty in its value, before and after data are collected and analyzed. P(y| ) is the probability of
y conditional on
) is the probability of
y conditional on
 , P(
, P( |y) is the probability of
|y) is the probability of  conditional on
y, P(y,
conditional on
y, P(y, ) is the
joint probability of the two variables, and P(
) is the
joint probability of the two variables, and P( ) (or [P(y]) is the
marginal probability of
) (or [P(y]) is the
marginal probability of  (or of
y), that is, the probability of
(or of
y), that is, the probability of  (or
y) regardless of the values of
y (or
(or
y) regardless of the values of
y (or
 ) .
) .
Three distributions of fundamental interest to Bayesians are included in this listing of probabilities.
P(
y|

): the probability distribution of the data (i.e., probability of the data), such as normal or gamma for continuous
y, and binomial, beta-binomial, Poisson, or negative binomial for discrete
y. Often called the
likelihood when calculated for specific observations. This distribution is the one used in classical frequentist analysis.
P(

): probability distribution of the parameter (or parameters) before the (new) data are collected. This is known as the
prior distribution.
P(

|
y): probability distribution of the parameter after the data are collected in an experiment or a survey. This is known as the
posterior distribution.
The joint probability can be written in several ways using rules of probability:
P(y, � ) = P(� ) = P(� ,
y) ,
y) | (1) |
P(y, � ) = P(� ) = P(� ǀ
y)·P(y) ǀ
y)·P(y) | (2) |
P(� ,
y) = P(yǀ � ,
y) = P(yǀ � ) ·P(� ) ·P(� ) ) | (3) |
Thus, equation (1) can be written as: P(

ǀ
y)·P(
y) = P(
yǀ

)·P(

), or
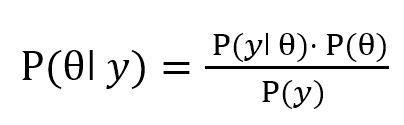 | (4) |
The denominator on the right-hand side in Equation 4, P(y), is based on integrating probabilities over all possible values of
 , and turns out to be a constant (although possibly
very difficult to calculate). Thus, Equation 4 is often written simply as a proportionality:
, and turns out to be a constant (although possibly
very difficult to calculate). Thus, Equation 4 is often written simply as a proportionality:
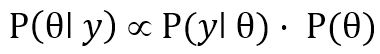
| (5) |
Equations 4 and 5 are typically known as the
Bayes’ formulae or
Bayes’ rules. Bayesian data analysis is all about estimating and interpreting the results, based on the data (y), the assumed distribution for
y, P(y| ), and the prior knowledge of (i.e., prior distribution of)
), and the prior knowledge of (i.e., prior distribution of)
 .
.
One should note that classical frequentist analysis is just based on P(y| ), without any consideration of prior distribution(s) for the parameter(s). In contrast, the essence of Bayesian analysis is to update the prior distribution based on the data in order to determine the posterior distribution of the parameter(s).
), without any consideration of prior distribution(s) for the parameter(s). In contrast, the essence of Bayesian analysis is to update the prior distribution based on the data in order to determine the posterior distribution of the parameter(s).
Prior distributions (or priors)
Prior distributions capture the quality, quantity and type of knowledge that one has about the parameters of interest (the ones to be estimated with the model and data). Basically, priors provide the evidence of what the parameters are, or look like, in the absence of “new” data. By “look”, we mean the range of values the parameters can take on, i.e., the dispersion (or spread) and skewness of the (possible) parameter values. The word ‘prior’ suggests a temporal relationship between parameters and data; however, this is not always true, because priors can be decided after data collection and implementation of Bayes’ rule.
Several priors or families of priors can be considered while conducting Bayesian analysis and a ‘correct’ prior does not exist in Bayesian analysis. This has been one of the points of arguments against Bayesian analysis. Priors are considerably important especially when data are few; however, their significance in the analysis will progressively diminish while the volume of data increases and most of the conclusions will rely on the “likelihood” function.
Priors are necessary in order to conduct Bayesian analysis. Not all elements of a prior distribution have to be known; some may be unknown and be specified in another layer of prior distributions known as
hyperprior distributions.
Types of prior distributions
i)
Noninformative prior (also known as
objective prior) indicates very little or no prior knowledge. This type of prior generally has minimum impact on the posterior distribution of a parameter of interest. The most common choice for a noninformative prior is a flat distribution, such as the uniform distribution, in the sense that assigns equal likelihood on all possible values of a parameter (Fig. 2) (Mila and Carriquiry 2004). Scientists often like noninformative priors because they seem more objective; however, they are not any more objective than other priors. Technically, flat
noninformative priors can create problems because they are not easy to construct and the model-fitting sampling algorithm may not converge well (see estimation discussion below). Thus, flat priors may produce unreliable results that are not suitable for inference.
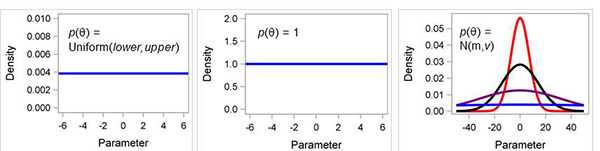
Figure 2. Examples of non-informative prior distributions. (A) uniform, (B) constant, or flat such as “1”, and (C) normal distribution N(m,v) where m is the mean and v is the variance; when variance is very large, ie., 106 then normal is similar to uniform (blue line).
Click to enlarge.
A very useful and practical choice for a
noninformative prior is the normal distribution with a large variance (Fig. 2C). Estimation methods often work well using this prior. A typical choice of a variance is 106, or larger. With such a large variance, the probability density is almost flat over a wide range of parameter values, thus closely mimicking a uniform distribution.
2)
Informative prior indicates prior knowledge of the parameter of interest, or assumed prior knowledge. This type of prior can be obtained from previous studies, historical data or elicitation from experts. An informative prior will have an impact on the posterior distribution of the parameter(s) and the degree of the impact will depend on the level of information in the prior and the amount of information available in the likelihood. Information on the likelihood is dependent on the number of observations in the data set. For instance, when a very strong informative prior is assigned to a likelihood consisting of few data points, it is expected that the prior will “dominate” in determining the posterior distribution (equation 4). On the other hand, when a strong informative prior is assigned to a likelihood consisting of a large number of data points, the prior will have a minimum impact on the posterior distribution of the parameter(s).
Many statistical distributions can be used for informative priors, but the normal distribution is the most common and a good practical choice for many circumstances (Fig. 3). Small variances restrict most of the density function to a narrow range. A straightforward way to link the variance to prior knowledge is to consider the range over which one believes the parameter lies. For example, if one is “95% confident” that the parameter ranges from 2.5 to 4.5, then the range is calculated as
range95 = 4.5-2.5 = 2. The variance of the normal prior is calculated as
v = (range95)2/16 or
v = 22/16 = 0.25 (Fig. 3).
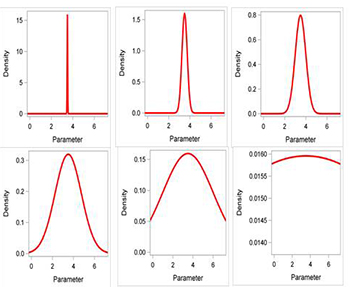
Figure 3. Normal distributions with increasing variances (0.000625, 0.0625, 0.25, 1.56, 6.25, 625.0).
Note the flattening of the curve with increasing variance.
Click to enlarge.
Posterior distributions (Equations 4 and 5) are typically difficult or impossible to calculate analytically because they require multidimensional integrations. The main challenge is in the inability to directly estimate P(y). There is a closed-form algebraic solution only for some very limited situations, and therefore, for many years, Bayesian applications were restricted to simple cases where so-called conjugate prior distributions were used.
The posterior distribution does not have a closed form for most models of relevance in plant pathology, such as generalized linear models, nonlinear models, and random-effects models. In such cases, exact inference is not possible. Fortunately, advances in computational methods, coupled with faster personal computers with large memories have drastically changed the possibilities for Bayesian analysis in recent years. Bayesian approaches can now be applied to most analytical problems in data analysis from a wide range of disciplines.
Summary
The relatively new Bayesian procedures available in SAS (e.g., GENMOD) provide a significant advantage to perform Bayesian analysis without great deal of computational skills. A ready set of priors and default samplers make Bayesian computing almost as easy as frequentist methods for many common problems in plant pathology. However, Bayesian analysis depends on the prior distributions, so it is important that the data analyst pays particular attention to the priors that are used and reports on the priors (whether the default or the user-selected non-informative or informative ones) in any paper or presentation. For more examples related to plant pathology, an extensive list of examples and SAS code can be found here:
http://oardc.osu.edu/APS-statsworkshop/downloads.htm (download Bayesian Workshop 2014.zip) The example file demonstrates the Bayesian approach to estimating a single mean and variance (one group problem), means for two groups (analogous to two-sample t-test), multiple group problem (one-way ANOVA analogy), and normal-distribution linear regression. More statistical and practical background on Bayesian analysis within GENMOD can be found in Stokes et al. (2014). For complex problems, one can use the more sophisticated MCMC procedure in SAS.
Literature Cited
Chen, F. 2009. Bayesian Modeling Using the MCMC Procedure in
Proceedings of the SAS Global Forum 2008 Conference, SAS Institute Inc., Cary, NC.
Gelman, A., Carlin, J.B, Stern, H.S., Dunson, D., Vehtari, A., and Rubin, D.B., 2013. Bayesian Data Analysis, 2nd edition, Chapman & Hall, CRC press.
Geweke, J. 1992. Evaluating the accuracy of sampling based approaches to calculating posterior moments, in Bayesian Statistics, V.4, J.M. Bernard, J.O. Berger, A.P. Dawiv, and F.M. Smith, eds. Clarendon Press, Oxford, UK.
Mila, A.L., and Carriquiry, A.L. 2004. Bayesian analysis in plant pathology. Phytopathology 94:1027-1030.
Mila, A.L., Driever, G.F., Morgan, D.P., and Michailides, T.J. 2005. Effects of latent infection, temperature, and precipitation pattern and irrigation on panicle and shoot blight progress in pistachio orchards. Phytopathology 95:926-932.
Mila, A.L., and Yuen, J. 2015. Use of Bayesian Methods. Pages 115-120 in Exercises in Plant Disease Epidemiology, K. L Stevenson and M. J. Jeger, eds. The American Phytopathological Society, APS Press, St. Paul, MN.
Spiegelhalter, D. J., Best, N. G., Carlin, B. P., and van der Linde, A. 2002. Bayesian measures of model complexity and fit (with discussion). J. of the Royal Stat. Soc. Series B 64:583-639.
Stokes, M., Chen, F., and Gunes, F. 2014. An introduction to Bayesian analysis with SAS/STAT software. Paper SAS400-2014. SAS, Inc.
https://support.sas.com/resources/papers/proceedings14/SAS400-2014.pdf
Additional Reading
Gelman, A. and Rubin, D.B. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7:457-472.
Heidelberger, P. and Welch, P.D. 1981. A spectral method for confidence interval generation and run length control of simulations. Communication of the ACM 24:233-245.
Heidelberger, P. and Welch, P.D. 1983. Simulation run length control in the presence of the initial transient. Operations research 31:11091144.
Raftery, A.E. and Lewis, S.M. 1992. One long run with diagnostics: Implementation strategies with Markov Chain Monte Carlo. Stat Sci. 7:493-497.
Raftery, A.E. and Lewis, S.M. 1996. The number of iterations, convergence diagnostics and generic Metropolis algorithms, in Markov Chain Monte Carlo in Practice, W.R. Gilks, D.J. Spiegelhalter, and S. Richardson, eds. Chapman & Hall, London, UK.
